Call the Doctor - HDF(5) Clinic
Table of Contents
- Clinic 2022-11-29
- Clinic 2022-10-04
- Clinic 2022-09-27
- Clinic 2022-08-30
- Clinic 2022-08-23
- Clinic 2022-08-16
- Clinic 2022-08-09
- Clinic 2022-08-02
- Clinic 2022-07-26
- Clinic 2022-07-19
- Clinic 2022-07-12
- Clinic 2022-07-05
- Clinic 2022-06-27
- Clinic 2022-06-21
- Clinic 2022-06-14
- Clinic 2022-06-07
- Clinic 2022-05-24
- Clinic 2022-05-17
- Clinic 2022-05-10
- Clinic 2022-04-26
- Clinic 2022-04-19
- Clinic 2022-04-12
- Clinic 2022-04-05
- Clinic 2022-03-29
- Clinic 2022-03-22
- Clinic 2022-03-15
- Clinic 2022-03-08
- Clinic 2022-03-01
- Clinic 2022-02-22
- Clinic 2022-02-15
- Clinic 2022-02-08
- Clinic 2022-02-01
- Clinic 2022-01-25
- Clinic 2022-01-18
- Clinic 2022-01-11
- Clinic 2022-01-04
- Clinic 2021-12-21
- Clinic 2021-12-07
- Clinic 2021-11-23
- Clinic 2021-11-16
- Clinic 2021-11-09
- Clinic 2021-11-02
- Clinic 2021-10-28
- Clinic 2021-10-19
- Clinic 2021-09-28
- Clinic 2021-09-21
- Clinic 2021-08-31
- Clinic 2021-08-24
- Clinic 2021-08-17
- Clinic 2021-08-10
- Clinic 2021-08-03
- Clinic 2021-07-27
- Clinic 2021-07-20
- Clinic 2021-07-13
- Clinic 2021-07-06
- Clinic 2021-06-29
- Clinic 2021-06-22
- Clinic 2021-06-15
- Clinic 2021-06-08
- Clinic 2021-06-01
- Clinic 2021-05-25
- Clinic 2021-05-18
- Clinic 2021-05-11
- Clinic 2021-05-04
- Clinic 2021-04-27
- Clinic 2021-04-20
- Clinic 2021-04-06
- Clinic 2021-03-30
- Clinic 2021-03-23
- Clinic 2021-03-16
- Clinic 2021-03-09
- Clinic 2021-03-02
- Clinic 2021-02-23
- Clinic 2021-02-09
- Goal(s)
- This is a meeting dedicated to your questions.
- In the unlikely event there aren't any
- Sometimes life deals you an HDF5 file
- Meeting Etiquette
- Be social, turn on your camera (if you've got one)
- Raise your hand to signal a contribution (question, comment)
- Be mindful of your "airtime"
- Introduce yourself
- Use the shared Google doc for questions and code snippets
- When the 30 min. timer runs out, this meeting is over.
- Notes
- Don't miss our next webinar about data virtualization with HDF5-UDF and how it can streamline your work
- Bug-of-the-Week Award (my candidate)
- Documentation update
- Clinic 2021-02-16
Clinic 2022-11-29
Your questions
- Q
- ???
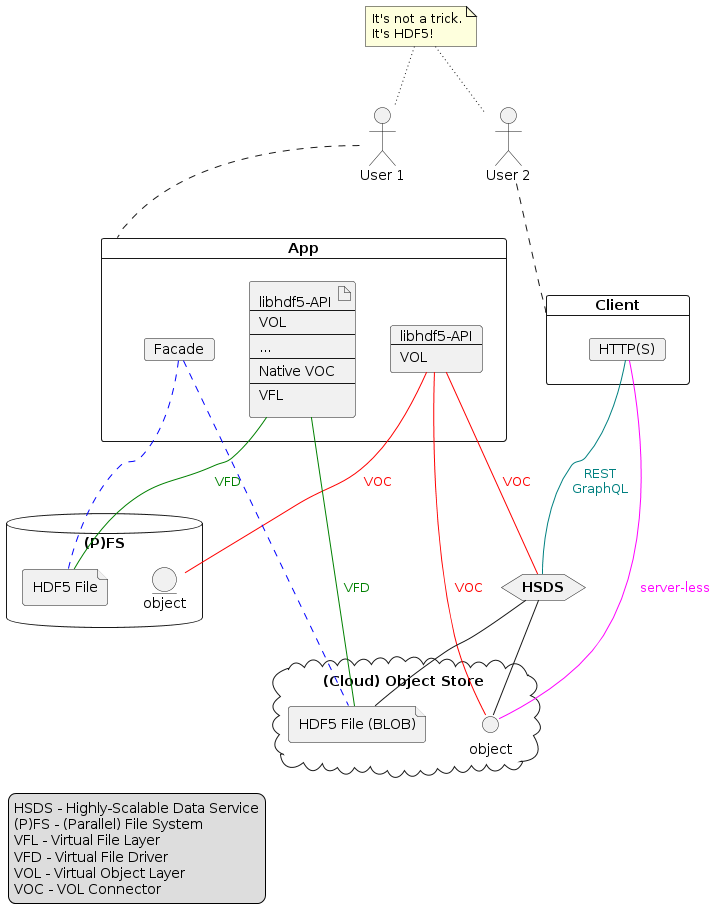
Types in the HDF5 Data Model
Reference: C.J. Date: Foundation for Object / Relational Databases: The Third Manifesto, 1998.
- Data Model
In the realm of HDF5, there are usually two data models in play.
- HDF5 Data Model
A data model is an abstract, self-contained, logical definition of the data structures, data operators, etc., that together make up the abstract machine with which users interact.
Metaphor: A data model in this sense is like a programming language, whose constructs can be used to solve many specific problems but in and of themselves have no direct connection with any such specific problem.
This is the sense in which I will be using the term 'data model.' I'm talking about HDF5 files, groups, datasets, attributes, transfers, etc.
An implementation of a given data model is a physical realization on a real machine of the components of the abstract machine that together constitute that model. There are currently two implementations of the HDF5 data model:
- HDF5 library + file format
- Highly Scalable Data Service (HSDS)
- Domain-specific Data Model
A data model is a model of the data (especially the persistent data) of some particular domain or enterprise.
Metaphor: A data model in this sense is like a specific program written in that language — it uses the facilities provided by the model, in the first sense of that term, to solve some specific problem.
We sometimes call this an HDF5 profile, the mapping of domain concepts to HDF5 data model primitives.
- HDF5 Data Model
- Types
In essence, a type is a set of (at least two) values — all possible values of some specific kind: for example, all possible integers, or all possible character strings, or all possible XML documents, or all possible relations with a certain heading (and so on).
(Some people require types to be named and finite.)
To define a type, we must:
- Specify the values that make up that type.
- Specify the hidden physical representation for values of that type. (This is an implementation issue, not a data model issue.)
- Specify a selector operator for selecting, or specifying, values of that type.
- Specify admissible type conversions and renderings.
- …
- Atomicity
The notion of atomicity has no absolute meaning; it just depends on what we want to do with the data. Sometimes we want to deal with an entire set of part numbers as a single thing, sometimes we want to deal with individual part numbers within that set—but then we’re descending to a lower level of detail, or lower level of abstraction.
- Scalar vs. Nonscalar Types
Loosely, a type is scalar if it has no user visible components and nonscalar otherwise. As atomicity, this has no absolute meaning. Some people treat 'scalar' and 'atomic' as synonymous.
- Values vs. Variables
Variables hold values. They have a location in space and time. Values "transcend" space and time.
- Questions
- What is the type of an HDF5 dataset?
- Is it a variable or a value?
- Is it scalar/nonscalar/atomic?
- What is an HDF5 datatype?
- What is the type of an HDF5 attribute?
- (How) Is an HDF5 user-defined function a dataset? (Extra credit!)
- Questions
- HDF5 Datatypes
The primary function of HDF5 datatypes is to describe the element type and physical layout of datasets, attributes, and maps. Documentation is a secondary function.
HDF5 supports a set of customizable basic types and a mechanism, datatype derivation, to compose new datatypes from existing ones. Any datatype derivation is rooted in atomic datatypes.
What's wrong with this figure? Nothing, but it's important to understand the context.
- What does 'Atomic' mean? (Not derived by composition. Composite = non-atomic = molecular)
- Why is an
Enumerationnot 'Atomic'? (Derives from a integer datatype.) - Why are
ArrayandVariable Lengthnot 'Atomic'? (Ditto. Derive from other datatype instances)
'Atomic' is an overloaded term in this context.
- Attribute values are treated as atomic under transfer by the HDF5 library
- Dataset values are not atomic because of partial I/O
- Selections
- Fields (for compounds)
- Dataset element values are atomic, except, records (values of compound)
Clinic 2022-10-04
Your questions
- Q
- ???
Last week's highlights
- Announcements
- Neil Fortner is our new Chief HDF5 Software Architect
- Happy days!
- Jobs @ The HDF Group
- Webinar Subfiling and Multiple dataset APIs: An introduction to two new features in HDF5 version 1.14
- Recording
- Slides: HDF5 Subfiling VFD & Multiple Dataset APIs
- Recent documentation updates
- Getting Started
- User Guide
- Reference Manual w/ language bindings (C, C++, Fortran, Java)
- Far from perfect but a big step forward
- Neil Fortner is our new Chief HDF5 Software Architect
- Forum
- C++ Thread unable to open file
- If you want threat-safety, use the C-API with a threat-safe build of the HDF5 library!
- Other language bindings (C++, Java, Fortran) and the high-level libs. are not thread-safe
- Performance improvements for many small datasets
- GraphQL is a bigger task
John provided an Python asynchronous I/O example
import asyncio import time async def say_after(delay, what): await asyncio.sleep(delay) print(what) async def main(): task1 = asyncio.create_task( say_after(2, 'hello')) task2 = asyncio.create_task( say_after(1, 'world')) print(f"started at {time.strftime('%X')}") # Wait until both tasks are completed (should take # around 2 seconds.) await task1 await task2 print(f"finished at {time.strftime('%X')}") asyncio.run(main())
started at 12:05:21 world hello finished at 12:05:23
- Noticeable speedup but not a panacea
Eventually I hope to have a version of
h5pydthat supportsasync(or maybe an entirely new package), that would make it a little easier to use. - Variable-Length Data in HDF5 Sketch RFC Status?
- Compound data type with zero-sized dimension
HDF5 array datatype field w/ a degenerate dimension
HDF5 "test.h5" { GROUP "/" { DATASET "dset" { DATATYPE H5T_COMPOUND { H5T_ARRAY { [2][2] H5T_IEEE_F32LE } "arr1"; H5T_ARRAY { [2][0] H5T_IEEE_F32LE } "arr2"; } DATASPACE SCALAR } } }- Use case?
- Metadata and Structure conventions
- ???
- C++ Thread unable to open file
- Engineering Corner
- Featuring: Dana Robinson
- What's new?
Starry HDF clinic line-up going forward
- 1st Tuesday of the month: Dana Robinson (HDF Engineering, Community)
- 2nd Tuesday: John Readey (HSDS, Cloud) – starting Oct 11
- 3rd Tuesday: Aleksandar Jelenak (Pythonic Science, Earth Science)
- 4th Tuesday: Scot Breitenfeld (HPC, Research)
- (In the unlikely event that there is a) 5th Tuesday: Gerd Heber + surprise guest
Clinic 2022-09-27
Your questions
- Q
- ???
Last week's highlights
- Announcements
- Thanks to Scot Breitenfeld, Aleksandar Jelenak, and John Readey for stepping up!
- Thanks to Lori the recordings are available on YouTube.
- Jobs @ The HDF Group
- Thanks to Scot Breitenfeld, Aleksandar Jelenak, and John Readey for stepping up!
- Forum
- Type confusion for chunk sizes
- Size matters
;-) - Use of
sizein APIs assize_t(C-type) orhsize_t(HDF5 API type) - What the
sizeunit?- Element count ->
hsize_t - Storage bytes ->
size_t
- Element count ->
- And we still managed to make a holy mess of it…
- Size matters
- Zstandard plugin for HDF5 does not allow negative compression levels
- Community life
- Store a group in a contiguous file space
Can one store all datasets in a group into a contiguous file space? I have an application that reads groups as units and all the datasets use the contiguous data layout. I believe this option, if available, can yield a good read performance.
- Interesting suggestions
- Elena suggested to first create all datasets with
H5D_ALLOC_TIME_LATE - Mark suggested to
H5Pget_meta_block_sizeto set aside sufficient "group space" (Maybe alsoH5Pset_est_link_info?)
- Elena suggested to first create all datasets with
- Interesting suggestions
- MSC Nastran *.H5
- HDFView "in distress" - it's not a substitute for a domain-specific viz tool
- What do people think of HDFView?
- Make HDFView available on Flathub.org
The app. has been accepted and is now available on flathub.org as org.hdfgroup.HDFView! That is HDFView 3.2.0 with HDF5 + HDF4 support for x86-64 & aarch64 CPUs.
- Let's try it!
- Performance improvements for many small datasets
- We just talked about multi-dataset I/O
- REST round-tripping
- GraphQL?
- pHDF,
H5Fflushand compression
- (Seemingly) Odd behavior of
H5Fflushin parallel with vs. w/o compression.
- (Seemingly) Odd behavior of
- Type confusion for chunk sizes
- Engineering Corner
- Featuring: Dana Robinson
- Is Dana back from his from trip?
Tips, tricks, & insights
Nada. (Just catching up on the forum…)
Clinic 2022-08-30
Your questions
- Q
- ???
Last week's highlights
- Announcements
- Webinar Vscode-h5web: A VSCode extension to explore and visualize HDF5 files
- August 31 (tomorrow) at 9:00 AM (CDT)
- Free registration
- Jobs @ The HDF Group
- New RFC
- Terminal VOL Connector Feature Flags
- How do I know if a VOL connector implements everything I need for my application?
- Webinar Vscode-h5web: A VSCode extension to explore and visualize HDF5 files
- Forum
Nothing stood out.
- Engineering Corner
- Featuring: Dana Robinson
- What do you want to see in “HDF5 2.0”?
- What else is new?
Tips, tricks, & insights
- Where is the "official" HDF5 standard?
- My colleague Aleksandar Jelenak just pointed me to HDF5 Data Model, File
Format and Library—HDF5 1.6
- An uncluttered and clear presentation
- Needs to be updated (e.g., VOL, HSDS) and revised, but still accurate
- The HDF Group maintains this HDF5 documentation
- My colleague Aleksandar Jelenak just pointed me to HDF5 Data Model, File
Format and Library—HDF5 1.6
- Proposed HDF Clinic format change
- Four themed events per month:
- Ecosystem (Aleksandar Jelenak)
- HDF5 & Python, R, Julia, other formats & frameworks, etc.
- HSDS (John Readey)
- HDF5 & Cloud, Kubernetes, server-less, etc.
- HPC (Scot Breitenfeld)
- Parallel I/O, file systems, DAOS, diagnostics, troubleshooting, etc.
- Engineering (Dana Robinson)
- Everything about getting involved and contributing to HDF5, PRs, issues, releases, new features, etc.
- We might have to adjust the weekday/time
- Thoughts?
- Four themed events per month:
Clinic 2022-08-23
Your questions
- Q
- ???
Last week's highlights
- Announcements
- Webinar Vscode-h5web: A VSCode extension to explore and visualize HDF5 files
- Aug 31, 2022 04:00 PM (Central) <– Really?
- Free Registration
- Jobs @ The HDF Group
- New blog post by Mr. HSDS (John Readey)
- HSDS Streaming
- Overcoming the
max_request_sizelimit - New performance numbers HSDS vs. HDF5 lib. and Mac Pro vs. HP Dev One
- Webinar Vscode-h5web: A VSCode extension to explore and visualize HDF5 files
- Forum
- Allocating chunks in a specific order?
- Use case: Zarr shards
- No API currently
- Potential issues:
- Relying on implementation details
- Undesirable constraints on library and tool behavior
- Performance impact
- Machine-readable units format?
- Blast from the past
- (Physical) Units are part of the datatype \(1.0 \mathrm{[m]} \neq 1.0 \mathrm{[s]}\)
- User-defined datatypes extend to user-defined units
- Datatype conversion pipeline extends to unit conversion pipeline
- Maybe time for another proposal attempt?
- C++ Read Compound dataset without taking static structure for different fieldnames
I have multiple H5 files and i am creating a generic code where i need to read specific field defined in input whether it will be an
arrayordoubleorint.- Great topic for tips and tricks…
- Allocating chunks in a specific order?
- Engineering Corner
- Featuring: Dana Robinson
- What do you want to see in “HDF5 2.0”?
- What else is new?
Tips, tricks, & insights
- HDF5 compound datatype introspection
- What's known at compile time?
- Field name(s) + in-memory type:
constexprortemplate - Field name(s) only: dataype introspection
- Field name(s) + in-memory type:
- Datatype introspection "algorithm":
- Retrieve the field in-file datatype via
H5Tget_member_indexandH5Tget_member_class/H5Tget_member_type - Retrieve the in-memory (or native) datatype via
H5Tget_native_type - Determine the size via
H5Tget_size - Construct an in-memory compound datatype via
H5Tcreate/H5Tinsert - Allocate a buffer of the right size
H5Dread& parse the buffer
- Retrieve the field in-file datatype via
- What's known at compile time?
Clinic 2022-08-16
Your questions
- Q
- ???
Last week's highlights
- Announcements
- HDF5 1.13.2 is out. Get it here!
- Highlights: Onion VFD, Subfiling VFD
- Dana?
- HDF5 1.13.2 is out. Get it here!
- Forum
- Collective VDS creation
We have data scattered across thousands of processes. But each process does not own an hyperslab of the data. It actually owns a somewhat random collection of pieces of data, which are all hyperslabs. To write this data to a file collectively, I first tried to make each process select a combination of hyperslabs using
H5Scombine_hyperslab.… Now I am trying to reproduce this behaviour using VDS: all pieces are written in a dataset in disorder. Another dataset (virtual this time) maps all pieces in the right order. It works when using 1 process!
The problem: I am unable to create this VDS in a collective way.
… -> is it possible to create a VDS collectively ?
H5Scombine_hyperslabis for point selections onlyNeil's answer RE: VDS
For the VDS question: to create a VDS collectively every process needs to add every mapping. This conforms with the way collective metadata writes in HDF5 generally work - every process is assumed to make exactly the same (metadata write) calls with exactly the same parameters. It would be possible to implement what you are describing as a high level routine which would call H5Pgetvirtual then do an allgather on the VDS mappings, but for now it’s easiest to handle it in the user application.
It’s also worth noting that there are some other limitations to parallel VDS I/O:
printfstyle mappings are not supported- The VDS must be opened collectively
- When using separate source file(s), the source file(s) cannot be opened by the library while the VDS is open.
- When using separate source file(s), data cannot be written through the VDS unless the mapping is equivalent to 1 process per source file
- All I/O is independent internally (possible performance penalty)
- Each rank does an independent open of each source file it accesses (possible performance penalty)
I should also note that VDS is not currently tested in the parallel regression test suite so there may be other issues.
- 9BN rows/sec + HDF5 support for all python datatypes
- Allocating chunks in a specific order?
- Q
- Is there a way to allocate (not necessarily write) unfiltered chunks in a specific order?
- A
- Mimic w/
H5Pset_external. Then what?
- Collective VDS creation
- Engineering Corner
- Featuring: Dana Robinson
- What do you want to see in “HDF5 2.0”?
- What else is new?
Tips, tricks, & ruminations
- Dealing w/ concurrency "in" HDF5: ZeroMQ
from datetime import datetime import h5py from time import sleep import zmq context = zmq.Context() socket = context.socket(zmq.SUB) socket.connect("tcp://localhost:5555") socket.setsockopt(zmq.SUBSCRIBE, b'camera_frame') sleep(2) with h5py.File('camera_data.hdf5', 'a') as file: now = str(datetime.now()) g = file.create_group(now) topic = socket.recv_string() frame = socket.recv_pyobj() x = frame.shape[0] y = frame.shape[1] z = frame.shape[2] dset = g.create_dataset('images', (x, y, z, 1), maxshape=(x, y, z, None)) dset[:, :, :, 0] = frame i=0 while True: i += 1 topic = socket.recv_string() frame = socket.recv_pyobj() dset.resize((x, y, z, i+1)) dset[:, :, :, i] = frame file.flush() print('Received frame number {}'.format(i)) if i == 50: break
- The HDF5 library as an application extension acts like a wall for threads
- Before that wall becomes more permeable, maybe client/server is the better approach?
- Let's have a forum discussion around a suitable HDF5 ZeroMQ protocol!
Clinic 2022-08-09
Your questions
- Q
- ???
Last week's highlights
- Announcements
- Webinar recording available now: The Hermes Buffer Organizer
- Cloud Storage Options for HDF5 - a new blog post by Mr. HSDS (John Readey)
- Upcoming HDF5 Training Workshop for intermediate and advanced users with
Scot Breitenfeld (The HDF Group)
- (Free) Registration deadline: Monday, August 15 2022
- Event date/time: Aug 31, 2022, 10:00 AM - 3:00 PM (America/Chicago)
- Note: The participation of foreign nationals is subject to certain restrictions. Check the website for details!
- Forum
- Backporting
H5Dchunk_iterto 1.12 and 1.10?
- Great contribution by Mark Kittisopikul
- Let's do more of this!
- Using HDF5 for file backed Array
Sample
// Describe the dimensions. This example would mimic a 3D regular grid // of size 10 x 20 x 30 with another 2D array of 60 x 80 at each grid point. std::array<size_t, 3> tupleDimensions = {10, 20, 30}; std::array<size_t, 2> componentDimensions = {60, 80}; // Instantiate the DataArray class as an array of floats using the file // /tmp/foo.hdf5" as the hdf5 file and “/data” as the internal hdf5 path // to the actual data DataArray data("/tmp/foo.hdf5", "/data", tupleDimensions, componentDimensions); // Now lets loop over the data for(size_t z = 0; z < 10; z++) for(size_t y = 0; y < 20; y++) for(size_t x = 0; x < 30; x++) { size_t index = // compute proper index to the tuple for(size_t pixel = 0; pixel < 80 * 60; pixel++) { data[index + pixel] = 0; } }
- Responses from Steven (Mr. H5CPP) and Rick (Mr. HDFql)
Sliding cursors are coming to HDFql
int count; HDFql::execute("SELECT FROM dset INTO SLIDING(5) CURSOR"); count = 0; // whenever cursor goes beyond last element, HDFql automatically retrieves // a new slice/subset thanks to an implicit hyperslab // (with start=[0,5,10,...], stride=5, count=1 and block=5) while(HDFql::cursorNext() == HDFql::Success) { if (*HDFql::cursorGetDouble() < 20) { count++; } } std::cout << "Count: " << count << std::endl;
H5P_set_filtercd_valuesParameter
- Start w/ the filter registry
- Filter maintainers are responsible, ultimately
- Backporting
- Engineering Corner
- Featuring: Dana Robinson
- What do you want to see in “HDF5 2.0”?
- What else is new?
Tips, tricks, & insights
- Datatype conversion - reloaded
- Q
- What if source and destination datatype are of different size?
- A
- We use a conversion buffer!
#include "hdf5.h" #include <assert.h> #include <stdint.h> #include <stdio.h> #include <stdlib.h> // our conversion function // HACK: in a production version, you would inspect SRC_ID and DST_ID, etc. herr_t B32toU64(hid_t src_id, hid_t dst_id, H5T_cdata_t *cdata, size_t nelmts, size_t buf_stride, size_t bkg_stride, void *buf, void *bkg, hid_t dxpl) { herr_t retval = EXIT_SUCCESS; switch (cdata->command) { case H5T_CONV_INIT: printf("Initializing conversion function...\n"); // do non-trivial initialization here break; case H5T_CONV_CONV: printf("Converting...\n"); uint32_t* p32 = (uint32_t*) buf; uint64_t* p64 = (uint64_t*) buf; // the conversion happens in-place // since we don't want to overwrite elements, we need to // shift/convert starting with the last element and work our // way to the beginning for (size_t i = nelmts; i > 0; --i) { p64[i-1] = (uint64_t) p32[i-1]; } break; case H5T_CONV_FREE: printf("Finalizing conversion function...\n"); // do non-trivial finalization here break; default: break; } return retval; } int main() { int retval = EXIT_SUCCESS; // in-memory bitfield datatype hid_t btfd = H5T_NATIVE_B32; // in-file unsigned integer datatype hid_t uint = H5T_STD_U64LE; // register our conversion function w/ the HDF5 library assert(H5Tregister(H5T_PERS_SOFT, "B32->U64", btfd, uint, &B32toU64) >= 0); // sample data uint32_t buf[32]; for (size_t i = 0; i < 32; ++i) { buf[i] = 1 << i; printf("%u\n", buf[i]); } hid_t file = H5Fcreate("foo.h5", H5F_ACC_TRUNC, H5P_DEFAULT, H5P_DEFAULT); hid_t fspace = H5Screate_simple(1, (hsize_t[]) {32}, NULL); hid_t dset = H5Dcreate(file, "integers", uint, fspace, H5P_DEFAULT, H5P_DEFAULT, H5P_DEFAULT); hid_t dxpl = H5Pcreate(H5P_DATASET_XFER); // we supply a conversion buffer where can convert up to 6 elements // at a time (48 = 6 x 8) unsigned char tconv[48]; H5Pset_buffer(dxpl, 48, tconv, NULL); // alternative: the HDF5 library will dynamically allocate conversion // and background buffers if we pass NULL for the buffers // H5Pset_buffer(dxpl, 48, NULL, NULL); // the datatype conversion function will be invoked as part of H5Dwrite H5Dwrite(dset, btfd, H5S_ALL, H5S_ALL, dxpl, (void*) buf); H5Pclose(dxpl); H5Dclose(dset); H5Sclose(fspace); H5Fclose(file); // housekeeping assert(H5Tunregister(H5T_PERS_SOFT, "B32->U64", btfd, uint, &B32toU64) >= 0); return retval; }
Clinic 2022-08-02
Your questions
- Q
On the tar to h5 converter: Does the (standard) compactor simply skip the >64K tar file entries if ones are encountered (w/ log or error message), or must they all conform to 64K limit? Also, what was the typical range on the number of files extracted/converted? –Robert
- A
- Currently, the code will skip entries >64K, because the underlying
H5Dcreatewill fail. (The logic and code can be much improved.) It's better to first runarchive_checker_64kand see if there are any size warnings. Theh5compactorandh5shredderwere successfully run on TAR archives with tens of millions of small images (<64K).
Last week's highlights
- Announcements
- Webinar recording "An Introduction to HDF5 for HPC Data Models, Analysis, and Performance" posted.
- Webinar Announcement: The Hermes Buffer Organizer
- Friday, August 5, 2022 at 11:00 a.m. Central Time
- Registration
- Webinar recording "An Introduction to HDF5 for HPC Data Models, Analysis, and Performance" posted.
- Forum
- Variable length list of strings
- What's a string?
- What the user meant: array of characters
- Ergo: list of variable-length strings
""= list of arrays of characters of varying length - This is not the same as an HDF5 dataset of variable-length strings
Aleksandar's example:
import h5py import numpy as np import string vlength = [3, 8, 6, 4] dt = h5py.vlen_dtype(np.dtype('S1')) with h5py.File('char-ragged-array.h5', 'w') as f: dset = f.create_dataset('ragged_char_array', shape=(len(vlength),), dtype=dt) for _ in range(len(vlength)): dset[_] = np.random.choice(list(string.ascii_lowercase), size=vlength[_]).astype('S1')
Sample output:
HDF5 "char-ragged-array.h5" { GROUP "/" { DATASET "ragged_char_array" { DATATYPE H5T_VLEN { H5T_STRING { STRSIZE 1; STRPAD H5T_STR_NULLPAD; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; }} DATASPACE SIMPLE { ( 4 ) / ( 4 ) } DATA { (0): ("g", "b", "x"), ("k", "j", "u", "u", "i", "p", "a", "t"), (2): ("t", "i", "b", "x", "u", "y"), ("q", "i", "r", "h") } } } }
- What's a string?
- Variable length list of strings
- Engineering Corner
- Featuring: Dana Robinson
- What do you want to see in “HDF5 2.0”?
- Releases on track?
Tips, tricks, & insights
- 9BN rows/sec + HDF5 support for all python datatypes
- Forum post
- Tablite GitHub repo
- Tutorial
Where is HDF5?
from tablite.config import H5_STORAGE H5_STORAGE
PosixPath('/tmp/tablite.hdf5')Simple schema: tables -> columns -> pages (via attributes)
/ Group /column Group /column/1 Dataset {NULL} /column/16 Dataset {NULL} ... /page Group /page/1 Dataset {3/Inf} /page/2 Dataset {3/Inf} ... /table Group /table/1 Dataset {NULL} /table/3 Dataset {NULL} ...
Clinic 2022-07-26
Corrections
- The VSCode extension for HDF5 works with NetCDF-4 files
- I grabbed a netCDF-3 file. Duh!
Your questions
- Q
- ???
Last week's highlights
- Announcements
- Webinar Announcement: An Introduction to HDF5 for HPC Data Models, Analysis, and Performance
- Presented by The HDF Group's one and only Scot Breitenfeld
- Final boarding call: Register here!
- VFD SWMR beta 2 release
- Testers wanted
- New blog post HSDS Docker Images by Mr. HSDS (John Readey)
- America runs on Dunkin, HSDS runs on Docker…
- DockerHub, container registries, Kubernetes,etc.
- Webinar Announcement: An Introduction to HDF5 for HPC Data Models, Analysis, and Performance
- Forum
- Looking for an hdf5 version compatible with go1.9.2
- Go HDF5!
;-)- Old software…
- Go HDF5!
- Looking for an hdf5 version compatible with go1.9.2
- Engineering Corner
- Featuring: Dana Robinson
- What do you want to see in “HDF5 2.0”?
- Evolution not revolution
- "Lessons learned" release
Tips, tricks, & insights
- Little HDF5 helpers for ML
- Use case: Tons of tiny image, audio, or video files
- Poor ML training I/O performance, especially against PFS
- Can't read w/o un-
tar-ing (uncompressing) - No parallel I/O
tar2h5- convert Tape ARchives to HDF5 files- Different optimizations
- Size optimization (compact datasets)
- Duplicate removal (SHA-1 checksum)
- Compression
- Different optimizations
- Use case: Tons of tiny image, audio, or video files
Clinic 2022-07-19
Your questions
- Q
- ???
Last week's highlights
- Announcements
- Webinar Announcement: An Introduction to HDF5 for HPC Data Models, Analysis, and Performance
- Presented by The HDF Group's one and only Scot Breitenfeld
- Register here!
- VFD SWMR beta 2 release
- Testers wanted
- VSCode extension for HDF5
- Brief demo later
- Webinar Announcement: An Introduction to HDF5 for HPC Data Models, Analysis, and Performance
- Forum
The week before last was quiet, but then it got hot…
ros3driver breaks randomly
- Not properly recovering from S3 read failures
- No retries
- On Python,
fsspecmight be the better option for the time being
- Not properly recovering from S3 read failures
- Memory mapping / Paging via HDF5 & VFD?
- Goes back to a HUG 2021 discussion
Maybe we don't need memory mapping API, but
What we really need is the ability to allocate space in the HDF5 file without writing. (@kittisopikulm)
- Interesting idea
- Possibility of backporting
H5Dchunk_iterto 1.12?
- 1.12 is nearing EOL
- 1.10 might be a better target
- HDFView on Windows not displaying fixed length UTF-8 string correctly
- Probably an HDFView bug
- Virtual datasets in HSDS?
- Not currently supported in HSDS, but…
- You can fake it via
H5D_CHUNKED_REF_INDIRECT
- You can fake it via
- Not currently supported in HSDS, but…
- implement hdf5 in eclipse cdt for a c tool and eclipse mingw 32 need help
- Are we failing developers?
Tips, tricks, & insights
- First look at the VSCode extension for HDF5
- If you don't have a local installation of VSCode, you can use GitHub Codespaces and run in a Web browser
- Or roll your own w/
code-serverand run in the browser
Clinic 2022-07-12
Your questions
- Q
- ???
Last week's highlights
- Announcements
- Deep Dive: HSDS Container Types
- New blog post by Mr. HSDS (John Readey)
- Webinar Announcement: An Introduction to HDF5 for HPC Data Models, Analysis, and Performance
- Presented by The HDF Group's one and only Scot Breitenfeld
- Register here!
- VFD SWMR beta 2 release
- Testers wanted
- Deep Dive: HSDS Container Types
- Forum
It was all quiet on the forum. What happened? Everybody must be happy, I guess…
Tips, tricks, & insights
- HDF5-style datatype conversion
- Use case: A lot of data coming from devices in physical experiments are
bitfields. To be useful for analysts, the data needs to be converted to
engineering types and units. Does that mean we always must store 2 or more
data copies?
- There is no intrinsic reason to have multiple copies, but there might be other reasons, e.g., performance, to maintain multiple copies.
- How do we pull this off? Filters? (No! Same datatype…)
- Datatype transformations!
- Example: Use an in-memory bitfield representation and store as a compound w/ two fields
Code#include "hdf5.h" #include <assert.h> #include <stdint.h> #include <stdio.h> #include <stdlib.h> // our compound destination type in memory typedef struct { uint16_t low; uint16_t high; } low_high_t; // our conversion function // HACK: in a production version, you would inspect SRC_ID and DST_ID, etc. herr_t bits2cmpd(hid_t src_id, hid_t dst_id, H5T_cdata_t *cdata, size_t nelmts, size_t buf_stride, size_t bkg_stride, void *buf, void *bkg, hid_t dxpl) { herr_t retval = EXIT_SUCCESS; switch (cdata->command) { case H5T_CONV_INIT: printf("Initializing conversion function...\n"); // do non-trivial initialization here break; case H5T_CONV_CONV: printf("Converting...\n"); // the conversion function simply splits and swaps bitfield halves low_high_t* ptr = (low_high_t*) buf; for (size_t i = 0; i < nelmts; ++i) { uint16_t swap = ptr[i].low; ptr[i].low = ptr[i].high; ptr[i].high = swap; } break; case H5T_CONV_FREE: printf("Finalizing conversion function...\n"); // do non-trivial finalization here break; default: break; } return retval; } int main() { int retval = EXIT_SUCCESS; // in-memory bitfield datatype hid_t btfd = H5T_NATIVE_B32; // compound of two unsigned shorts hid_t cmpd = H5Tcreate(H5T_COMPOUND, 4); H5Tinsert(cmpd, "low", 0, H5T_NATIVE_USHORT); H5Tinsert(cmpd, "high", 2, H5T_NATIVE_USHORT); // register our conversion function w/ the HDF5 library assert(H5Tregister(H5T_PERS_SOFT, "bitfield->compound", btfd, cmpd, &bits2cmpd) >= 0); // notice that the conversion function is its own inverse assert(H5Tregister(H5T_PERS_SOFT, "compound->bitfield", cmpd, btfd, &bits2cmpd) >= 0); // sample data uint32_t buf[32]; for (size_t i = 0; i < 32; ++i) { buf[i] = 1 << i; printf("%ud\n", buf[i]); } // we could check the conversion in-memory, in-place //H5Tconvert(btfd, cmpd, 32, buf, NULL, H5P_DEFAULT); hid_t file = H5Fcreate("cmpd.h5", H5F_ACC_TRUNC, H5P_DEFAULT, H5P_DEFAULT); hid_t fspace = H5Screate_simple(1, (hsize_t[]) {32}, NULL); hid_t dset = H5Dcreate(file, "shorties", cmpd, fspace, H5P_DEFAULT, H5P_DEFAULT, H5P_DEFAULT); // the datatype conversion function will be invoked as part of H5Dwrite H5Dwrite(dset, btfd, H5S_ALL, H5S_ALL, H5P_DEFAULT, (void*) buf); // the datatype conversion function will be invoked as part of H5Dread H5Dread(dset, btfd, H5S_ALL, H5S_ALL, H5P_DEFAULT, (void*) buf); for (size_t i = 0; i < 32; ++i) printf("%ud\n", buf[i]); H5Dclose(dset); H5Sclose(fspace); H5Fclose(file); // housekeeping assert(H5Tunregister(H5T_PERS_SOFT, "compound->bitfield", cmpd, btfd, &bits2cmpd) >= 0); assert(H5Tunregister(H5T_PERS_SOFT, "bitfield->compound", btfd, cmpd, &bits2cmpd) >= 0); H5Tclose(cmpd); return retval; }
- Datatype conversions do what it says on the tin and sometimes are just what you need
- Don't confuse them with filters!
- With HDF5-UDF it is possible to implement datatype conversions via filters, but at the expense of additional datasets
- Use case: A lot of data coming from devices in physical experiments are
bitfields. To be useful for analysts, the data needs to be converted to
engineering types and units. Does that mean we always must store 2 or more
data copies?
Clinic 2022-07-05
Your questions
- Q
- ???
Last week's highlights
- Announcements
- VFD SWMR beta 2 release
- Testers wanted
- VFD SWMR beta 2 release
- Forum
- HDF5 1.10, 1.12 dramatic drop of performance vs 1.8
- Recap:
h5open_f/h5close_foverhead, chunk size 1 issue… User provided a nice table
chunk size data (bytes) metadata (bytes) 1 120000 1128952 50 120000 26456 2048 122880 3400 131072 524288 3400 - No compression
- Steven Varga suggested that
- The application might benefit from a dedicated I/O handler, and he provided a multithreaded queue example
- Data generationn can be decoupled from recording via ZeroMQ, and he provided an example with the generator (sender) in Fortran and the recorder (receiver) in C
- Recap:
- Issue w/ memory backed files
- We are able to reproduce the issue!
- Slowdown in
H5Oget_infobecause ofH5O_INFO_ALLoption - The difference between the in-file and in-memory image sizes can be explained by incremental (re-)allocation in the core VFD
The real issue is this:
HDF5-DIAG: Error detected in HDF5 (1.13.2-1) thread 0: #000: H5F.c line 837 in H5Fopen(): unable to synchronously open file major: File accessibility minor: Unable to open file #001: H5F.c line 797 in H5F__open_api_common(): unable to open file major: File accessibility minor: Unable to open file #002: H5VLcallback.c line 3686 in H5VL_file_open(): open failed major: Virtual Object Layer minor: Can't open object #003: H5VLcallback.c line 3498 in H5VL__file_open(): open failed major: Virtual Object Layer minor: Can't open object #004: H5VLnative_file.c line 128 in H5VL__native_file_open(): unable to open file major: File accessibility minor: Unable to open file #005: H5Fint.c line 1964 in H5F_open(): unable to read superblock major: File accessibility minor: Read failed #006: H5Fsuper.c line 450 in H5F__super_read(): unable to load superblock major: File accessibility minor: Unable to protect metadata #007: H5AC.c line 1396 in H5AC_protect(): H5C_protect() failed major: Object cache minor: Unable to protect metadata #008: H5C.c line 2368 in H5C_protect(): can't load entry major: Object cache minor: Unable to load metadata into cache #009: H5C.c line 7315 in H5C__load_entry(): incorrect metadata checksum after all read attempts major: Object cache minor: Read failed
- It appears that the superblock checksum (in versions 2, 3) is not correctly set or updated
- Maybe this is a corner case of the use of HDF5 file image w/ core VFD? Investigating…
- Authenticated AWS S3
- Use of the read-only S3 VFD w/ Python?
The ros3 driver is very slick. It works great for public h5 files.
I am trying to perform a read of an H5 file using ros3 that requires authentication credentials. I have been successful in using h5ls/h5dump in accessing an h5 file against an authenticated AWS S3 call using –vfd=ros3 and –s3-cred=(<region,<keyid>,<keysecret>).
However, I cannot get this to work with h5py.
- The user answered his on question,
h5pydoes support this VFD and AWS credentials as documented here - He also commented that this is not limited to AWS S3, but also works with other S3-compatible storage options
- And he's looking for HSDS on GCP (Google Cloud)
- HDF5 1.10, 1.12 dramatic drop of performance vs 1.8
Tips, tricks, & insights
- What others are doing with HDF5
- Subscribe for alerts on Google Scholar
- Alert query
hdf5- …
- Type
- Most relevant results
- You will get 2-4 emails a week with about 6-10 citations
- Recent highlights
- Fission Matrix Processing Using the MCNP6.3 HDF5 Restart File
- See the Wikipedia article for Monte Carlo N-Particle Transport Code
Towards A Practical Provenance Framework for Scientific Data on HPC Systems
So we derive an I/O-centric provenance model, which enriches the W3C PROV standard with a variety of concrete sub-classes to describe both the data and the associated I/O operations and execution environments precisely with extensibility.
Moreover, based on the unique provenance model, we are building a practical prototype which includes three major components: (1) Provenance Tracking for capturing diverse I/O operations; (2) Provenance Storage for persisting the captured provenance information as standard RDF triples; (3) User Engine for querying and visualizing provenance.
Stimulus: Accelerate Data Management for Scientific AI applications in HPC
… a lack of support for scientific data formats in AI frameworks. We need a cohesive mechanism to effectively integrate at scale complex scientific data formats such as HDF5, PnetCDF, ADIOS2, GNCF, and Silo into popular AI frameworks such as TensorFlow, PyTorch, and Caffe. To this end, we designed Stimulus, a data management library for ingesting scientific data effectively into the popular AI frameworks. We utilize the StimOps functions along with StimPack abstraction to enable the integration of scientific data formats with any AI framework. The evaluations show that Stimulus outperforms several large-scale applications with different use-cases such as Cosmic Tagger (consuming HDF5 dataset in PyTorch), Distributed FFN (consuming HDF5 dataset in TensorFlow), and CosmoFlow (converting HDF5 into TFRecord and then consuming that in TensorFlow) by 5.3x, 2.9x, and 1.9x respectively with ideal I/O scalability up to 768 GPUs on the Summit supercomputer. Through Stimulus, we can portably extend existing popular AI frameworks to cohesively support any complex scientific data format and efficiently scale the applications on large-scale supercomputers.
-
DIARITSup is a chain of various software following the concept of ”system of systems”. It interconnects hardware and software layers dedicated to in-situ monitoring of structures or critical components. It embeds data assimilation capabilities combined with specific Physical or Statistical models like inverse thermal and/or mechanical ones up to the predictive ones. It aims at extracting and providing key parameters of interest for decision making tools. Its framework natively integrates data collection from local sources but also from external systems. DIARITSup is a milestone in our roadmap for SHM Digital Twins research framework. Furthermore, it intends providing some useful information for maintenance operations not only for surveyed targets but also for deployed sensors.
Meanwhile, a recorder manage the recording of all data and metadata in the Hierarchical Data Format (HDF5). HDF5 is used to its full potential with its Single-Writer-Multiple-Readers feature that enables a graphical user interface to represent the saved data in real-time, or the live computation of SHM Digital Twins models for example. Furthermore, the flexibility of HDF5 data storage allows the recording of various type of sensors such as punctual sensors or full field ones.
- It's impossible to keep up, but always a great source of inspiration!
- Fission Matrix Processing Using the MCNP6.3 HDF5 Restart File
- Subscribe for alerts on Google Scholar
- Next time: datatype conversions
Clinic 2022-06-27
Your questions
- Q
- ???
Last week's highlights
- Announcements
- VFD SWMR beta 2 release
- Testers wanted
- 2022 HDF5 Release Schedule announced
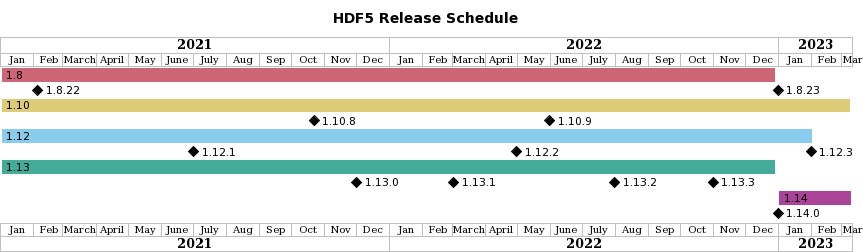
- HDF5 1.8.x and 1.12.x are coming to an end (1.8.23, 1.12.3)
- HDF5 1.10.x and 1.14.x are here to stay for a while
- Performance work over the Summer
- HDF5 1.13.2: Selection I/O, VFD SWMR, Onion VFD (late July/early August)
- HDF5 1.13.3: Multi-dataset I/O, Subfiling (late September/early October)
- HDF5 1.14.0 in late December/early January
- We are hiring
- Director of Software Engineering
- Chief HDF5 Software Architect
- Interested in working for The HDF Group? Send us your resume!
- Speed up cloud access using multiprocessing! by John Readey (Mr. HSDS)
Each file is slightly more than 1 TB in size, so downloading the entire collection would take around a month with a 80 Mbit/s connection. Instead, let’s suppose we need to retrieve the data for just one location index, but for the entire time range 2007-2014. How long will this take?
- VFD SWMR beta 2 release
- Forum
- How efficient is HDF5 for data retrieval as opposed to data storage?
I would like to a keyed 500GB table into HDF5, and then then retrieve rows matching specific keys.
For an HDF5 file, items like all the data access uses an integer “row” number, so seems like I would have to implement a 'key to row number map" outside of HDF5.
Isn’t retrieval more efficient with a distributed system like Spark which uses HDFS?
- Too open-ended question w/o additional context
- There are no querying or indexing capabilities built into HDF5
- Can something like this be implemented on top of HDF5? It's been done many times, for specific requirements.
- Are other tools more efficient? Maybe. Maybe not.
- HDF5 1.10, 1.12 dramatic drop of performance vs 1.8
Profiling (gperftools) revealed
85% of the time is spent in
H5Fget_obj_count.- The user correctly inferred that this is related to frequent
h5open_f/h5close_f, and an issue we covered in the past- Bug in
h5f_get_obj_count_f& fixed in PR#1657 by Scot.
- Bug in
- After applying that fix performance is on par w/ 1.8
Happy ending! (almost)
h5stat output: ... Summary of file space information: File metadata: 1201344 bytes Raw data: 99260 bytes Amount/Percent of tracked free space: 0 bytes/0.0% Unaccounted space: 17392 bytes Total space: 1317996 bytes
- Greater than 1:10 data to metadata ratio
- Usually means trouble
- Culprit: chunk size of 1
- Greater than 1:10 data to metadata ratio
- Issue w/ memory backed files
- Still puzzled
- How efficient is HDF5 for data retrieval as opposed to data storage?
Tips, tricks, & insights
- SWMR (old) and compression "issues" - take 2
- Free-space management is disabled in the original SWMR implementation
- Can lead to file bloat when compression is enabled & overly aggressive flushing
- Question: How does the new SWMR VFD implementation behave?
- Untested, but should work in principle
- There is an updated VFD SWMR RFC
Clinic 2022-06-21
Your questions
- Q
- ???
Last week's highlights
- Announcements
- VFD SWMR beta 2 release
- Testers wanted
- HDFView 3.1.4 released
- Some confusion around versioning
- HDFView 3.2.x series is based on the HDF5 1.12.x releases.
- HDFView 3.1.x series is based on the HDF5 1.10.x releases.
- HDFView 3.3.x series will be based on the future 1.14.x releases.
- Known issue in 3.2.0: HDFView crashes on an attribute of VLEN of REFERENCE
- Some confusion around versioning
- VFD SWMR beta 2 release
- Forum
- Check if two identifiers refer to the same object
H5Imodule- Identifiers are transient
- Pre-defined and user-defined identifier types
- Pre-defined IDs identify HDF5 objects but also VFL and VOL plugins, etc.
- See
H5I_type_t
- See
- User-defined ID types must be first registered w/ the library before use
Certain functions are only available for user-defined IDs
[1] H5Iobject_verify: Object atom/Unable to find ID group information cannot call public function on library type
- Assuming pre-defined HDF5 IDs for objects, use
H5Oget_infoto retrieve a structure that contains an address or token which can then be compared
- Issue w/ memory backed files
- Workflow:
- Create a memory-backed file (w/ core VFD)
- Make changes
- Get a file image (in memory) and share w/ another process (shared mem. or network)
- Create another memory-backed file from this file image
- Should work just fine w/
H5P[g,s]et_file_image
- Workflow:
- Multithread Writing to two files
- Some users don't know how to help themselves.
- Check if two identifiers refer to the same object
Tips, tricks, & insights
- SWMR (old) and compression "issues"
- Reports:
- Elena answered in both cases
- It's not a bug but a "feature"
- File space recycling is disabled in SWMR mode
- Every time a dataset chunk is flushed, new space is allocated in the file
- How to alleviate?
- Keep the chunk in cache & flush only once it's baked
- Don't compress edge chunks
- Bigger picture: We need a feature (in-)compatibility matrix! Long overdue!
- Question: How does the new SWMR implementation behave?
Clinic 2022-06-14
Your questions
- Q
- ???
Last week's highlights
- Announcements
- Hermes 0.7.0-beta release
- Bug fixes
- Google OR tools replaced by GLPK
- VFD SWMR beta 2 release
- Testers wanted
- HUG 2022 Europe videos available
- Agenda and YouTube playlist
- Thanks to Lori Cooper
- Hermes 0.7.0-beta release
- Forum
- Multithread Writing to two files
Reading Image data from single file and writing to two h5 files using threads. Getting run time error doing this. What could be the reason ?
- It turns out the user is using high-level library (
hdf5_hl.h) functions, e.g.,H5IMmake_image_24bit - These functions are not thread-safe, even if the underlying HDF5 library was built w/ thread-safety enabled
- Workaround: Use the thread-safe core library and follow the specification, such as HDF5 Image and Palette Specification Version 1.2
- It turns out the user is using high-level library (
H5Rdestroywhen stored
If a reference is stored as an attribute to an object, should
H5Rdestroystill be called?- Yes
- Fake it till you make it: GANs with HDF5
- Never the same HDF5 dataset (value) twice!
- A clever combination of HDF5-UDF and generative adversarial networks (GAN)
- Competition between two neural networks a (synthetic-) data generator and a discriminator
- TensorFlow Keras API supports storing trained model weights in HDF5 files
- We can collocate the HDF5-UDF to synthesize data with the generator model
- Reading from this dataset creates a new sample every time
- Multithread Writing to two files
Tips, tricks, & insights
- HDF5 command-line tools and HDF5 path names w/ special characters
- Example:
/equilibrium/time_slice[]&profiles_2d[]&theta_SHAPE - We have a problem!
h5ls
First try
gerd@penguin:~$ h5ls equilibrium.h5/equilibrium/time_slice[]&profiles_2d[]&theta_SHAPE [2] 8959 [3] 8960 -bash: profiles_2d[]: command not found -bash: theta_SHAPE: command not found [3]+ Exit 127 profiles_2d[] gerd@penguin:~$ time_slice[]**NOT FOUND** ^C [2]+ Exit 1 h5ls equilibrium.h5/equilibrium/time_slice[] gerd@penguin:~$
Double quotes to the rescue
gerd@penguin:~$ h5ls "equilibrium.h5/equilibrium/time_slice[]&profiles_2d[]&theta_SHAPE" time_slice[]&profiles_2d[]&theta_SHAPE Dataset {107/Inf, 3/Inf, 2/Inf}
h5dump
First attempt
gerd@penguin:~$ h5dump -pH -d "/equilibrium/vacuum_toroidal_field&b0" equilibrium.h5 HDF5 "equilibrium.h5" { DATASET "/equilibrium/vacuum_toroidal_field&b0" { DATATYPE H5T_IEEE_F64LE DATASPACE SIMPLE { ( 107 ) / ( H5S_UNLIMITED ) } STORAGE_LAYOUT { CHUNKED ( 107 ) SIZE 656 (1.305:1 COMPRESSION) } FILTERS { COMPRESSION DEFLATE { LEVEL 1 } } FILLVALUE { FILL_TIME H5D_FILL_TIME_IFSET VALUE -9e+40 } ALLOCATION_TIME { H5D_ALLOC_TIME_INCR } } }Second attempt
gerd@penguin:~$ ~/packages/bin/h5dump -pH -d "/equilibrium/time_slice[]&profiles_2d[]&z_SHAPE" equilibrium.h5 HDF5 "equilibrium.h5" {h5dump error: unable to get link info from "/equilibrium/time_slice[]&profiles_2d"}
Not good. Need to investigate.
- Example:
Clinic 2022-06-07
Your questions
- Q
- ???
Last week's highlights
- Announcements
- Release of HDF5-1.10.9
- Parallel compression improvements
- Doxygen-based User Guide draft
- HDF5 is now tested and supported on macOS 11.6 M1
- VS 2015 will no longer be supported starting in 2023
- VFD SWMR beta 2 release
- Testers wanted
- 2022 European HDF5 Users Group
- Was a HUGE success
- Thanks to Lana Abadie of ITER, Andy Gotz of ESRF, and Lori Cooper of HDFG
- Stay tuned for the presentation videos!
- They will be posted on the YouTube channel
- Release of HDF5-1.10.9
- Forum
- Make HDFView available on Flathub.org
There's been some progress
Mostly working except filesystem portal integration, which makes the entire thing pretty useless for now, the code may need patching…
- Not sure what that means
- HDF5 write perf - Regular API vs Direct Chunk on uncompressed dataset
- Samuel posted
pprofprofiles- Need time to digest
- Samuel posted
- Make HDFView available on Flathub.org
Tips, tricks, & insights
- I can't create a compound datatype with XXX (too many) fields
- Cause: size limitation of the datatype message
- Possible solutions:
- Break up the compound type into multiple compound types
- Use a group and make each field a dataset
- Use an opaque type and store metadata to parse
- Use multiple (extendible) 2D datasets
- One dataset for each field datatype
- Keep the field metadata (names, order) in attributes
Clinic 2022-05-24
Your questions
- Q
- Element order preservation in point selections?
- Q
- ???
Last week's highlights
- Announcements
- 2022 European HDF5 Users Group (HUGE)
- HDF5 1.10.9-2-rc-1 source available for testing
- Parallel Compression improvements
- First steps toward a Doxygen-based “User Guide”
- HDF5 is now tested and supported on macOS 11.6 M1
- 2022 European HDF5 Users Group (HUGE)
- Forum
- Survey on the usage of self-describing data formats
- A minor step towards thread concurrency
- Idea
- Drop the global lock (temporarily) whenever we are performing file I/O
- Interesting idea, but devil's in the details (memory/file-hybrid state)
- Make HDFView available on
flathub.org
- Again, very interesting idea
- How can we help?
- VSCode extension for HDF5
- Someone started it already. Duh!
- How "complete" is it? (What are the requirements?)
- How can it be improved?
- We'll discuss this at HUG Europe 2022
- HDF5 write perf - Regular API vs Direct Chunk on uncompressed dataset
Some good news: no performance regression from HDF5 1.8.22
- direct chunk hyperslab chunk hyperslab contiguous regular 19.060877 38.609770 23.500788 never fill 19.578496 19.223011 24.316146 latest fmt 19.697542 35.719180 24.503973 never fill + latest format 19.084984 18.970671 24.817945 HDF5 1.12.1
- direct chunk hyperslab chunk hyperslab contiguous regular 21.050749 35.889814 23.982538 never fill 18.917586 20.929941 23.291541 latest fmt 19.921759 35.472971 23.935622 never fill + latest format 19.646342 19.800098 24.406450 - Question remains: Why is
hyperslab contiguousconsistently 25% slower than the chunky versions?- Need to dig deeper
- Read the code or … (both)
- gperftools (
pprof) output visualized w/[q,k]cachegrind
- Need to dig deeper
- Survey on the usage of self-describing data formats
Tips, tricks, & insights
- Awkward arrays
- Project home
- Awkward Array: Manipulating JSON-like Data with NumPy-like Idioms
- SciPy 2020 presentation by Jim Pivarski
- Watch this!
How would you represent something like this in HDF5? (Example from Jim's video)
import awkward as ak array = ak.Array([ [{"x": 1, "y": [11]}, {"x": 4, "y": [12, 22]}, {"x": 9, "y": [13, 23, 33]}], [], [{"x": 16, "y": [14, 24, 34, 44]}] ])
- Compound? Meh! Empty or partial records? Variable-length sequences…
=:-O - Columnar layout
- Pick an iteration order
- Put record fields into contiguous buffers
Keep track list offsets
outer offsets: 0, 3, 3, 5 content for x: 1, 4, 9, 16 offsets for y: 0, 1, 3, 6, 10 content for y: 11, 12, 22, 13, 23, 33, 14, 24, 34, 44
- A picture is worth a thousand words (screenshot from Jim's presentation)
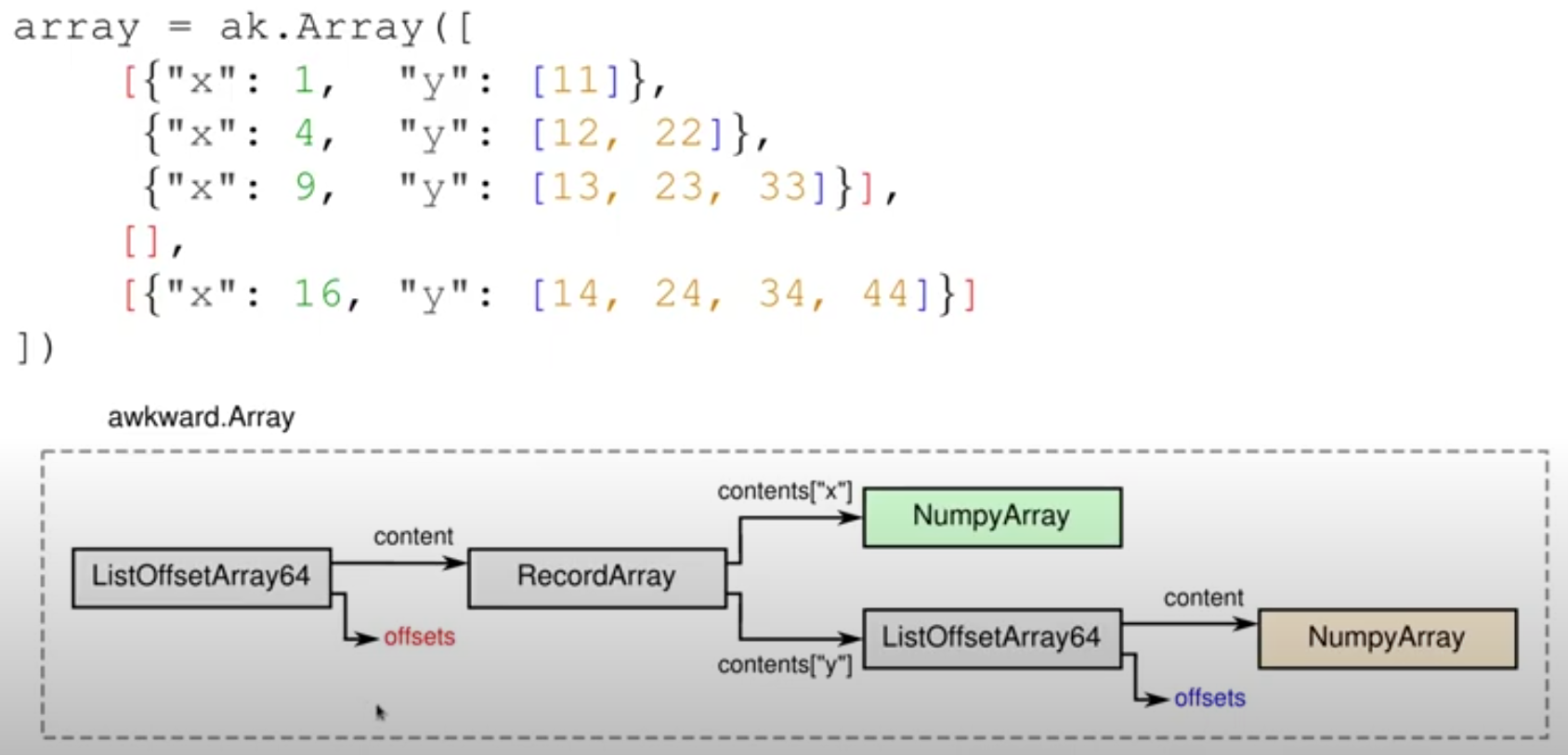
- See the HDF5 structure?
- You can store both iteration orders, if that's what you need
- See the HDF5 structure?
- Compound? Meh! Empty or partial records? Variable-length sequences…
- Food for thought: Sometimes HDF5 is about finding balance between appearance and performance
- We will have a few HEP (High-Energy Physics)-themed presentations at HUG
Europe 2022
- You have been warned
;-)
- You have been warned
Clinic 2022-05-17
Your questions
- Q
- Element order preservation in point selections?
- Q
- ???
Last week's highlights
- Announcements
- 2022 European HDF5 Users Group (HUGE)
- Release of HDFView 3.2.0
- SWMR support (configurable refresh timer)
- Display and edit attribute data in table views, the same way as datasets
- Read and display non-standard floating-point numbers
- HDF5 VOL Status Report – Exascale Computing Project
- Includes early DAOS VOL connector performance numbers
- HSDS Data Streaming Arrives
- 100 MB (configurable) cap on HTTP request size
- Otherwise
413 - Payload Too Largeerror - Increasing the
max_request_sizecan help only if HSDS Docker container or Kubernetes pod has sufficient RAM - Now what? Streaming to the rescue!
- Let clients see bytes returning from the server while that is still processing the tail chunks in the selection
- Otherwise
- 100 MB (configurable) cap on HTTP request size
- 2022 European HDF5 Users Group (HUGE)
- Forum
- HDF5 write perf - Regular API vs Direct Chunk on uncompressed dataset
- We provided a little test program to eliminate as many layers as possible
The user ran the tests and obtained these results (seconds via
clock_gettime(CLOCK_PROCESS_CPUTIME_ID,.)):- direct chunk hyperslab chunk hyperslab contiguous regular 1.631005 3.368258 2.069583 never fill 1.632858 1.641345 2.069205 latest fmt 1.643421 3.233817 2.068250 never fill + latest format 1.633859 1.611976 2.029573 - Why is
hyperslab contiguousconsistently 25% slower than the chunky versions?- Perhaps some extra buffer copying going on here??? TBD
- Failures with insufficient disk space
- The HDF5 library state appears to be inconsistent after a disk-full error
(
ENOSPC)- What's the state of a file in that situation? (Undefined)
- Recovering to the last sane state is harder than it may seem, if not impossible
- But things appear to be worse: The library crashes (assertion failure) on
shutdown
- Open handle accounting is screwed up
- What's the state of a file in that situation? (Undefined)
- See the reproducer in last week's notes!
- It appears that the problem might be Windows-specific.
- @Dana tried to reproduce the problem under Linux and couldn't
- Nobody has come forward so far with a non-Windows error report
- The HDF5 library state appears to be inconsistent after a disk-full error
(
- Corrupted file due to shutdown
Thank you for all your help and all of the very useful pointers. I managed to recover the data in the file. I did not get all the names of the datasets but with the mentioned attribute i was able to reconstruct the data.
Thank you and all the best, Christian
- Great!
- A lost business opportunity
;-), but also a sign that skill, determination, and documentation go a long way - Our way always was and will be: Open-Source Software and Specifications
- HDF5 write perf - Regular API vs Direct Chunk on uncompressed dataset
Tips, tricks, & insights
Next time…
Clinic 2022-05-10
Your questions
- Q
- ???
Last week's highlights
- Announcements
- 2022 European HDF5 Users Group (HUGE)
- Hermes 0.6.0-beta release
- Highlight: Hermes HDF5 Virtual File Driver
- GitHub
- HDF5 1.12.2 is out
- Parallel compression improvements
- Support for macOS Apple M1 11.6 Darwin 20.6.0 arm64 with Apple clang version 12.0.5.
- The shortened versions of the long options (ex:
--datasinstead of--dataset) have been removed from all the tools.
- Sharing Experience on HDF5 VOL Connectors Development and Maintenance
- Wednesday, May 11, 1:00-2:00 PM ET
- Part of the ECP 2022 Community BOF days
- Open registration
- 2022 European HDF5 Users Group (HUGE)
- Forum
- Select mulptiple hyperslabs in some order
- (Hyperslab) Selections can be combined via set-theoretic operations (union, intersection, …)
- There's an implicit (C or Fortran) ordering of the selected grid points, i.e., (within commutativity rules of set theory) the order of those operations doesn't matter
- What about point selections?
hdf5vshffile
- Naming conventions (extensions) for HDF5 files
- Technically, it doesn't matter
- Tools, such as HDFView, use search filters, e.g.,
*.h5and*.hdf5
- HDF5 write perf - Regular API vs Direct Chunk on uncompressed dataset
h5pyhaswrite_directandwrite_direct_chunk- About a 50% difference in performance. Why?
- What could slow down
H5Dwrite(vsH5Dwrite_chunk) when having no data conversion, no data scattering (contiguous layout), no chunking, no filters?
- Failures with insufficient disk space
- The HDF5 library state appears to be inconsistent after a disk-full error
(
ENOSPC)- What's the state of a file in that situation? (Undefined)
- Recovering to the last sane state is harder than it may seem, if not impossible
- But things appear to be worse: The library crashes (assertion failure) on
shutdown
- Open handle accounting is screwed up
- What's the state of a file in that situation? (Undefined)
Reproducer
#include "hdf5.h" #include <stdint.h> #include <stdlib.h> #define SIZE (1024 * 1024 * 128) int work(const char* path) { int retval = EXIT_SUCCESS; uint8_t* data = (uint8_t*) malloc(sizeof(uint8_t)*SIZE); for (size_t i = 0; i < SIZE; ++i) { *(data+i) = i % 256; } hid_t fapl = H5I_INVALID_HID; if ((fapl = H5Pcreate(H5P_FILE_ACCESS)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_fapl; } if (H5Pset_fclose_degree(fapl, H5F_CLOSE_STRONG) < 0) { retval = EXIT_FAILURE; goto fail_file; } hid_t file = H5I_INVALID_HID; if ((file = H5Fcreate(path, H5F_ACC_TRUNC, H5P_DEFAULT, fapl)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_file; } hid_t group = H5I_INVALID_HID; if ((group = H5Gcreate(file, "H5::Group", H5P_DEFAULT, H5P_DEFAULT, H5P_DEFAULT)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_group; } hid_t fspace = H5I_INVALID_HID; if ((fspace = H5Screate_simple(1, (hsize_t[]) {(hsize_t) SIZE}, NULL)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_fspace; } hid_t dset = H5I_INVALID_HID; if ((dset = H5Dcreate(group, "H5::Dataset", H5T_NATIVE_UINT8, fspace, H5P_DEFAULT, H5P_DEFAULT, H5P_DEFAULT)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_dset; } if (H5Dwrite(dset, H5T_NATIVE_UINT8, fspace, fspace, H5P_DEFAULT, data) < 0) { retval = EXIT_FAILURE; goto fail_write; } printf("Write succeeded.\n"); if (H5Fflush(file, H5F_SCOPE_GLOBAL) < 0) { retval = EXIT_FAILURE; goto fail_flush; } printf("Flush succeeded.\n"); fail_flush: fail_write: if (H5Dclose(dset) < 0) { printf("H5Dclose failed.\n"); } fail_dset: if (H5Sclose(fspace) < 0) { printf("H5Sclose failed.\n"); } fail_fspace: if (H5Gclose(group) < 0) { printf("H5Gclose failed.\n"); } fail_group: if (H5Fclose(file) < 0) { printf("H5Fclose failed.\n"); } fail_file: if (H5Pclose(fapl) < 0) { printf("H5Pclose failed.\n"); } fail_fapl: return retval; } int main() { int retval = EXIT_SUCCESS; retval &= work("O:/foo.h5"); // limited available space to force failure retval &= work("D:/foo.h5"); // lots of free space return retval; }
- The HDF5 library state appears to be inconsistent after a disk-full error
(
- External datasets relative to current directory or HDF5 file as documented?
- Documentation error: Thanks to the user who reported it.
- Select mulptiple hyperslabs in some order
Tips, tricks, & insights
Next time…
Clinic 2022-04-26
Your questions
- Q
- ???
Last week's highlights
- Announcements
- Forum
- Bug? H5TCopy of empty enum
- Is an empty enumeration datatype legit?
- I couldn't find that ticket @Elena mentioned a while ago
- Where is it?
- Bug? H5TCopy of empty enum
Tips, tricks, & insights
- Documentation - variable-length datatype description seems incomplete (doesn't mention global heap)
Interesting GitHub issue
I'm working on trying to decode an HDF5 file (attached) manually using the specification (for the purpose of writing rust code for a decoder that quickly extracts raw signal data from nanopore FAST5 files)…
- Tools of the trade
h5dump,h5debug,h5check - Highlights gaps/inaccuracies/inconveniences in the file format specification
- Bit-level representation of variable-length string datatypes
- What's a parent type? (spec. doesn't say)
- What's the parent type of a VLEN string? (character)
- (How) Is it encoded in the datatype message?
- Use of the global heap for VLEN data
- Not discussed where VLEN datatype message is discussed
- Bit-level representation of variable-length string datatypes
Sample
HDF5 "perfect_guppy_3.6.0_LAST_gci_vs_Nb_mtDNA.fast5" { GROUP "/" { ATTRIBUTE "file_version" { DATATYPE H5T_STRING { STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_UTF8; CTYPE H5T_C_S1; } DATASPACE SCALAR DATA { (0): "2.0" } } GROUP "read_44dcef85-283e-4782-b7b1-1c9a0f682597" { ATTRIBUTE "run_id" { DATATYPE H5T_STRING { STRSIZE H5T_VARIABLE; STRPAD H5T_STR_NULLTERM; CSET H5T_CSET_ASCII; CTYPE H5T_C_S1; } DATASPACE SCALAR DATA { (0): "cb45e5bda47a362d52bcfa146df9083b463bf65e" } } ...2.0\0is322e 3000in hex.- Let's find it!
Clinic 2022-04-19
Your questions
- Q
- ???
Last week's highlights
- Announcements
- 2022 European HDF5 Users Group (HUG)
- HDF5 1.12.2-3-rc-1 source available for testing
- Parallel compression improvements backported from HDF5 1.13.1
- HSDS v0.7beta13
- Support for Fancy Indexing
- Unlike with h5py though, HSDS works well with long lists of indexes by parallelizing access across the chunks in the selection.
- For example, retrieving 4000 random columns from a 17,520 by 2,018,392
dataset demonstrated good scaling as the number of HSDS nodes was increased:
- 4 nodes: 65.4s
- 8 nodes: 35.7s
- 16 nodes: 23.4 s
- The limit on the number of chunks accesses per request has been removed
- Support for Fancy Indexing
- 2022 European HDF5 Users Group (HUG)
- Forum
- Help Request regarding Java 3.3.2
- Switched from HDF Java 2.6.1 to 3.3.2 & seeing errors not seen before
- Artifact of checking for HDF5 Image convention attributes
- Prior versions ignored exceptions silently
8-( - Now we have "expected errors"???
h5f_get_obj_count_fExtremely Buggy?
- User is trying to use
h5fget_obj_count_fto establish the number of open handles; it appears that the result depends on the number of timesh5open_fwas called (???) - Calling
H5openw/ the C-API is usually not necessary and multiple calls have no side-effects h5open_f(Fortran API) appears to behave differently, buggy?- Fixed by Scot Breitenfeld in HDFFV-11306 Fixed #1657
- What a turnaround!
- User is trying to use
- C++ Read h5 cmpd (/struct) dataset that each field is vector
User's data
typedef struct { std::vector FieldA, FieldB, FieldC; } TestH5; TestH5.FieldA = {1.0,2.0,3.0,4.0}; // similar to FieldB, FieldC
- How to read this
TestH5using C++?
This brings us to today's …
- Help Request regarding Java 3.3.2
Tips, tricks, & insights
- How H5CPP makes you ask the right questions
(All quotations from Steven Varga's response!)
The expression below is a [templated] Class datatype in C++, placed in a non-contiguous memory location, requiring scatter-gather operators and a mechanism to dis-assemble reassemble the components. Because of the complexity AFAIK there is no automatic support for this sort of operation.
template <typename T> struct TestH5{ std::vector<T> FieldA, FieldB, FieldC; };
The structure above maybe modelled in HDF5 in the following way:
- (Columnar)
/group/[fieldA, fieldB, fieldC]fast indexing by columns, more complex and slower indexing by rows; also easier read/write from Julia/Python/R/C/ etc… - (Records) by a vector of tuples:
std::vector<std::tuple<T,T,T>>where you work with a single dataset, fast indexing by rows and slower indexing by columns - (Blocked) exotic custom solution based on direct chunk write/read: fast indexing of blocks by row and column wise at the increased complexity of the code.
- (Hybrid) …
H5CPP provides mechanism for the first two solutions:
TestH5<int> data = {std::vector<int>{1,2,3,4}, std::vector<int>{5,6,7}, std::vector<int>{8,9,10}}; h5::fd_t fd = h5::create("example.h5",H5F_ACC_TRUNC); h5::write(fd, "/some_path/fieldA", data.fieldA); h5::write(fd, "/some_path/fieldB", data.fieldB); h5::write(fd, "/some_path/fieldC", data.fieldC);
Ok the above is simple and well behaved, the second solution needs a POD struct backing, as tuples are not supported in the current H5CPP version (the upcoming will support arbitrary STL)
struct my_t { int fieldA; int fieldB; int fieldC; }
You can have any data types and arbitrary combination in the POD struct, as long as it qualifies as POD type in C++. This approach involves H5CPP LLVM based compiler assisted reflection – long word, I know; sorry about that. The bottom line you need the type descriptor and this compiler does it for you, without lifting a pinky.
std::vector<my_t> data; h5::fd_t fd = h5::create("example.h5",H5F_ACC_TRUNC); h5::write(fd, "some_path/some_name", data);
This approach is often used in event recorders, hence there is this
h5::appendoperator to help you out:h5::ds_t ds = h5::open(...); for(const auto& event: event_provider) h5::append(ds, event);
Both of the layouts are used to model sparse matrices, the second resembling COO or coordinate of points, whereas the first is for Compressed Sparse Row|Column format.
Slides are here, the examples are here.
best wishes: steve
Go on and read the rest of the thread! There is a lot of good information there (layouts, strings, array types, …).
- (Columnar)
Clinic 2022-04-12
Your questions
- Q
- ???
Last week's highlights
- Announcements
- Forum
- What kind of STL containers do you use in your field?
- Does anybody use the STL?
;-)
- Does anybody use the STL?
- Bug? H5TCopy of empty enum
- Looks like one
- Broken HDF5 file cannot be opened
- Don't make a fuzz!
;-) - Cornering the HDF5 library by feeding it random byte sequences as attribute and link names (or values)
- Mission accomplished!
- Don't make a fuzz!
- Automated formatted display of STL-like containers
g++ -I./include -o pprint-test.o -std=c++17 -DFMT_HEADER_ONLY -c pprint-test.cpp g++ pprint-test.o -lhdf5 -lz -ldl -lm -o pprint-test ./pprint-test LISTS/VECTORS/SETS: --------------------------------------------------------------------- array<string,7>:[xSs,wc,gu,Ssi,Sx,pzb,OY] vector:[CDi,PUs,zpf,Hm,teO,XG,bu,QZs] deque:[256,233,23,89,128,268,69,278,130] list:[95,284,24,124,49,40,200,108,281,251,57, ...] forward_list:[147,76,81,193,44] set:[V,f,szy,v] unordered_set:[2.59989,1.86124,2.93324,1.78615,2.43869,2.04857,1.69145] multiset:[3,5,12,21,23,28,30,30] unordered_multiset:[gZ,rb,Dt,Q,Ark,dW,Ez,wmE,GwF] ADAPTORS: --------------------------------------------------------------------- stack<T,vector<T>>:[172,252,181,11] stack<T,deque<T>>:[54,278,66,70,230,44,121,15,58,149,224, ...] stack<T,list<T>>:[251,82,278,86,66,40,278,45,211,225,271, ...] priority_queue:[zdbUzd,tTknDw,qorxgk,mCcEay,gDeJ,FYPOEd,CIhMU] queue<T,deque<T>>:[bVG,Bbs,vchuT,FfxEw,CXFrr,JAx,sVlcI] queue<T,list<T>>:[ARPl,dddmHT,mEiCJ,OVEYS,FIJi,jbQwb,tpJnpj,rlCRoKn,nBKjJ,KPlU,jatsUI, ...] ASSOCIATE CONTAINERS: --------------------------------------------------------------------- map<string,int>:[{LID:2},{U:2},{Xr:1},{e:2},{esU:1},{kbj:1},{qFc:3}] map<short,list<string>>:[{LjwUkey:5},{jZxhk:6},{sxKKVu:8},{vSmHmu:8},{wRBTdGS:7}] multimap<short,list<int>>:[{ALpPkqbJ:[6,6,8,7,8,5,7,8,5,5,6, ...]},{AwsHR:[8,5,6,6,5,6,7,6,7,6,8, ...]},{HtLQMvHv:[5,7,6,7,8,6,7]},{KbseLYEs:[5,8,6,8]},{RzsJm:[7,6,8,7,7,7,7,6,6,8,7, ...]},{XpNSkhDa:[7,5,8,8,7,8,5,5,5]},{cXPImNk:[6,8,6,5,8,7,5,6,6,8,6, ...]},{gkKHyh:[5,8,6,6,6,6,5,5,6]},{iPmaraP:[7,6,7,6,7,7,5,7,5,7,7, ...]},{pLmqL:[6,5,5,5,6]}] unordered_map<short,list<string>>:[{udXahPXD:7},{hUgYjak:5},{OpOmaBqA:7},{vTldeWdS:5},{jEHQST:8},{UZxId:7},{IslGsnGY:8}] unordered_multimap<short,list<int>>:[{JldxFw:[5,6,8,6,6]},{tnzhP:[8,6,8,5,5,8,8,8]},{cvMaS:[5,7,5,5,5,5]},{eGlyp:[8,7,8,8,7]}] RAGGED ARRAYS: --------------------------------------------------------------------- vector<vector<string>>:[[pkwZZ,lBqsR,cmKt,PDjaS,Zj],[Nr,jj,xe,uC,bixzV],[uBAU,pXCa,fZEH,FIAIO],[Vczda,HKEzO,ySqr,Fjd,nh,pgb,zcsw],[fLCgg,qQ,Reul,aTXp,DENn,ZDtkV,VXcB]] array<vector<short>,N>:[[29,49,29,42,25,33,49,33,44,49],[50,48,35,22,35,33,33],[46,27,23,20,48,38,45,28,45],[25,33,41,22,36]] array<array<short,M>,N>:[[90,35,99],[47,58,53],[82,25,72],[76,92,62],[39,88,32]] MISC: --------------------------------------------------------------------- pair<int,string>:{6:hJnCm} pair<string,vector<int>>:{iLclgkjnoY:[2,5,6,6,7,3,7,3,3,4,8, ...]} tuple<string,int,float,short>:<XTK,3,2.63601,6> tuple<string,list<int>,vector<float>,short>:<[TaPryDWKv,attpFqqIc,geHwbX,vdZ,kvruDeaxpZ,dSOqbVpr,jTciLPgBbI,duc,yUZiCP,zGrTsweTk,LNouX, ...],4,5.81555,[2,3,5,6,8]> - Group Overhead - File size problem for hierarchical data
We found that a group has quite a memory overhead (was it 2 or 20 kb?).
- How was that measured?
- What's in a group?
- Responses from Steven, John, and GH
- The figure quoted looks unusual/too high
H5Pset_est_link_inforegression in 1.13.1?
- Yup, looks like it!
- Compact vs. dense group storage
- The documentation has a few clues
- Corrupted file due to shutdown
I have a problem with a HDF5 file created with h5py. I have the suspicion that the process writing to the file was killed.
Try common household tools
strings -t d file.h5
- Then reach for the heavier guns (
h5check)
- What kind of STL containers do you use in your field?
Clinic 2022-04-05
Your questions
- Q
- ???
Last week's highlights
- Announcements
- Forum
- File remains open after calling
HD5F.close
- Context: .NET,
HDF5.PInvokemigrating from 1.8.x to 1.10.x - Biggest change: switch to 64-bit handles
- Symptoms: Change in unit test behavior, file closure
- User forgot to switch from
inttolongin one place C-style
typedefhas no equivalent in C#; hack:#if HDF5_VER1_10 using hid_t = System.Int64; #else using hid_t = System.Int32; #endif
- How to discover open handles?
H5Fget_obj_count,H5Fget_obj_idsH5Iiterate(since HDF5 1.12)
- Context: .NET,
- File remains open after calling
Tips, tricks, & insights
Clinic 2022-03-29
Your questions
- Q
- ???
Last week's highlights
- Announcements
- Free seminar: Using HDF5 and HSDS for Open Science Research at Scale
- Friday, April 1, 2022, 11:00 am - 12:30 pm EDT
- 2022 European HDF5 Users Group (HUG)
- Free seminar: Using HDF5 and HSDS for Open Science Research at Scale
- Forum
- Error: Read failed
- Got a sample, nothing unusual
- Core VFD works just fine
- Need to find a way to reproduce the behavior (I/O kernel)
- Mystery continues
- Symbolic links with relative path
- What's the meaning of 'relative' in HDF5? W.r.t. handle (file, group, …)!
..does and cannot mean what it does in a shell (no tree structure)- Special characters in link names? (
/,.,./,/./) ..,..., etc., and spaces are legitimate link names
- Refresh object so that external changes were applied
- The state of an open HDF5 file is a hybrid (in-memory, in-file)
- Simultaneously opening an HDF5 file in different processes with one or more
writers, without inter-process communication, creates a coherence problem!
(and potentially a consistency problem…)
- Don't go there!
- Use SWMR, IPC, or I/O request queue!
- Read Portion of Dataset
- That's what HDF5 is all about
- Great responses from a number of people
- Most comprehensive example by Steven (H5CPP)
- Error: Read failed
Tips, tricks, & insights
Clinic 2022-03-22
Your questions
- Q
- ???
Last week's highlights
- Announcements
- Free seminar: Using HDF5 and HSDS for Open Science Research at Scale
- Friday, April 1, 2022, 11:00 am - 12:30 pm EDT
- 2022 European HDF5 Users Group (HUG)
- Free seminar: Using HDF5 and HSDS for Open Science Research at Scale
- Forum
- Check if Hard/Soft/External link is broken
- How to detect broken soft or external links?
H5Lexistschecks only for link existence in a group, not validity of destination
- Not a perfect match, but
H5Oexists_by_namemight do the trick
- How to detect broken soft or external links?
- A problem when saving
NATIVE_LDOUBLEvariables
- Interesting discussion around
NATIVE_LDOUBLE - Compiler and processor dependent => you are opening a can of worms
- Two options:
- User-defined FP datatype + soft conversions (relatively safe)
- Opaque datatype + metadata (dangerous)
- Interesting discussion around
- Error: Read failed
- Mystery continues
- Trying to get a sample
- Use the core VFD to check if it's potentially a POSIX VFD/Windows issue
- Reading and Writing Multiple Files Using Multiple Threads C++
- The HDF5 library can be built thread-safe
- Multiple threads can open HDF5 files independently
- BUT, currently, only a single thread can be executing code in the HDF5 library, i.e., there's no concurrency, at the moment
- Two options:
- (Intermediate) Have a I/O request queue and a single thread servicing I/O requests
- (Advanced) Carefully plan dataset allocation and file metadata discovery to do concurrent writes and reads outside the HDF5 library
- Memory management in
H5FD_class_t
- GitHub PR#1502
- Good discussion
- Check if Hard/Soft/External link is broken
Tips, tricks, & insights
(Out of time…)
Clinic 2022-03-15
Your questions
Re: split/multi driver question from Session 55. On review, just would like to confirm:
- Q
- Once the member files (
-s.h5, -b.h5, …) are written with theMULTIdriver (most likely on a POSIX filesystem), they must remain co-located. - A
- No, however care must be used when re-opening them.
- Q
- They can, however, be copied into a (read-only) S3 bucket.
- A
- Yes.
- Q
- An application can read the S3 multi HDF5 "file" by configuring both
the
MULTIfile driver and theROS3driver - A
- Yes, but this is untested.
- Q
- There is no provision for putting the raw data
-r.h5on S3 and the others (-s.h5, -b.h5, -g.h5, -l.h5, -o.h5) on a local POSIX filesystem. - A
- This should be possible, but has not been tested, AFAIK.
Last week's highlights
- Announcements
- Forum
- Reading multiple hdf5 files mounted on remote folder
- (Blast from the past)
- Not sure if that's an
h5pyissue
- A problem when saving
NATIVE_LDOUBLEvariables
- Wikipedia:long double
- Need more information
- Reading and Writing Multiple Files Using Multiple Threads C++
- Steven posted a good comment
- Too many options w/o additional context
- Reading multiple hdf5 files mounted on remote folder
Tips, tricks, & insights
(Out of time…)
Clinic 2022-03-08
Your Questions
- Q
- ???
Last week's highlights
- Announcements
- HDF5 1.13.1 release
- Release notes
- Blog post Parallel compression improvements in HDF5 1.13.1
- Bug fixes & performance improvements and better support for collective I/O
- Much improved IOR results from
Coriat NERSC - For parallel dataset creation early allocation was the only option
- Incremental file space allocation is now supported for datasets created in parallel but only if they have filters applied to them
- HDF5 1.13.1 release
- Forum
- Extent the dataspace of an existing attribute
- Storing multiple physical quantities in a single scalar dataset, e.g.,
velocity, density and temperature of a cubic with dimensional elements of
101 x 71 x 51, the dataset would have for example an extent of(3, 101, 71, 51) - How to keep track of quantity metadata, e.g., unit symbol?
- What to do if we add a (e.g., fourth) quantity to the dataset?
- Idea
Store the quantity metadata in an array-like attribute
- Problems
H5T_ARRAYclass datatypes are fixed rank and extent- Attribute dataspaces are not extendable
- Solutions
All solutions come with different trade-offs!
- Assuming this doesn't happen too often: extend/re-write the attribute on each dimension change
- Use a variable-length sequence (VLEN) datatype: read/re-write the attribute on each dimension change
Use a "shared" attribute (=attribute that is an object reference to an extendable dataset)
- Note
- For the use case at hand, the sharing aspect is not important. It's the extendability of the object-referenced dataset.
Figure 1: A "shared" attribute
- Storing multiple physical quantities in a single scalar dataset, e.g.,
velocity, density and temperature of a cubic with dimensional elements of
- Memory Leak while Reading Data?
- Loop over a directory of HDF5 files and populate a list of per-file dictionaries
- A bit of a mystery; appears to happen only under macOS
- No issues under Windows w/
h5py2.x or 3.x, according to the user
- No issues under Windows w/
- Extent the dataspace of an existing attribute
Tips, tricks, & insights
(Out of time…)
Clinic 2022-03-01
Your Questions
- Q
- ???
Last week's highlights
- Announcements
- Tutorial Announcement: Constructing a Simple Terminal VOL Connector
- Recording and slides available
- VOL toolkit on GitHub
- Tutorial Announcement: Constructing a Simple Terminal VOL Connector
- Forum
- Can Compound Dataset been compressed?
- Yes.
- Compound datatype with array datatype fields
- Misunderstanding on chunking and elements
- No Minimal Reproducible Example (MRE), no error messages
- Hdf5 crosscompile to arm64
- Memory management in
H5FD_class_t
- When the VFL layer was created (~1998?) there were only RAM and HDDs
- With GPU, FPGA, and other near-data computing devices there are new types of memory, storage, and storage access paths
- The (de-)allocation VFD callbacks deal with (de-)allocation of (virtual) file space
- I/O acceleration depends on device-specific memory
- Avoid reams of device specific conditonally-compiled allocation code
- Since the application selects a device-specific VFD, extend the
H5FD_class_tinterface with generic device-memory (de-)allocation callbacks My colleague Jordan Henderson recently added a control callback and a corresponding op-code.
/* * Defining H5FD_FEAT_MEMMANAGE for a VFL driver means that * the driver uses special memory management routines or wishes * to do memory management in a specific manner. Therefore, HDF5 * should request that the driver handle any memory management * operations when appropriate. */ #define H5FD_FEAT_MEMMANAGE 0x00010000 herr_t (*ctl)(H5FD_t *file, uint64_t op_code, uint64_t flags, const void *input, void **output);
- Can Compound Dataset been compressed?
Tips, tricks, & insights
- HDF5 extension APIs - Virtual File Layer (VFL)
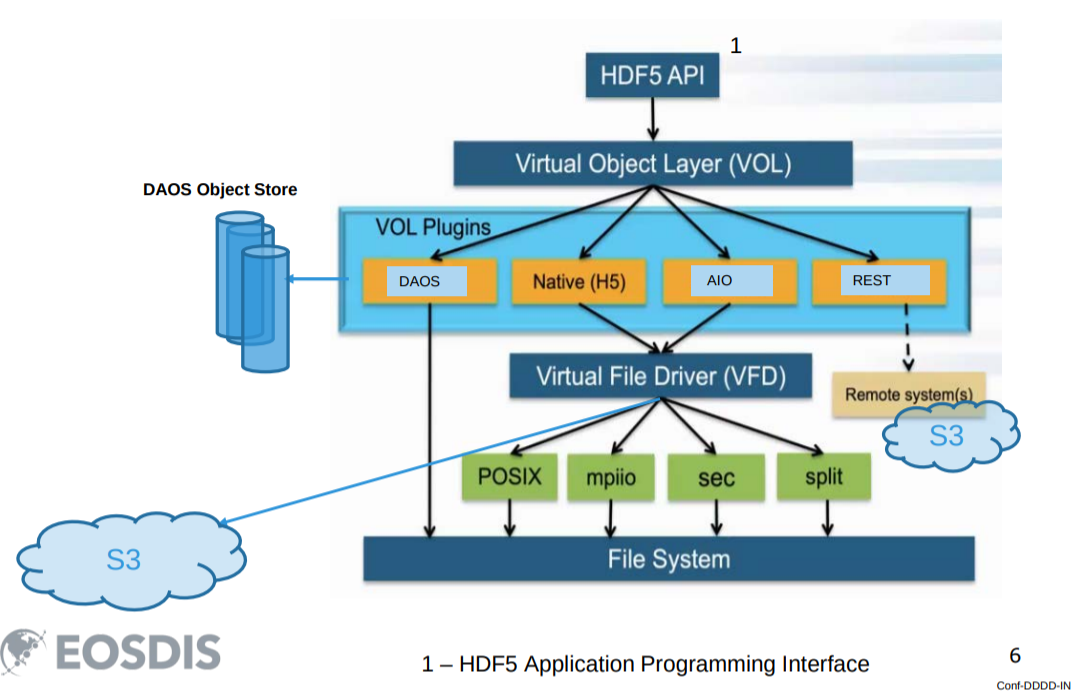
Figure 2: HDF5 extension APIs (Elena Pourmal, The HDF Group)
- No VFL toolkit, but using an existing VFD (= VFL plugin) is a good starting
point, see files called
H5FD*.[c,h]in the HDF5 source tree. - Example: read-only VFD plugin for Hadoop File System (HDFS)
- No VFL toolkit, but using an existing VFD (= VFL plugin) is a good starting
point, see files called
Clinic 2022-02-22
Your Questions
- Q
- Clarification/explanation on John Readey’s "seismic data" post regarding using native HDF5 as an HSDS "single object". –Robert Seip
Last week's highlights
- Announcements
- Constructing a Simple Terminal VOL Connector
- It's happening on this Friday, February 25th, 11:00 a.m. to 1:00 p.m. (Central)
- You can still register!
- Check out another interesting blog post
- A Kind of Magic: Storing Computations in HDF5
- The latest and greatest on Lucas C. Villa Real's HDF5-UDF
- A Kind of Magic: Storing Computations in HDF5
- Constructing a Simple Terminal VOL Connector
- Forum
- Unable to open a Dataset in my HDF5 file
- It often helps to read what people are actually asking…
I'm glad my colleague Aleksandar did just that and clarified that the way to obtain a NumPy array from a dataset is
phrase_numpy = phrase[...]and not
phrase_numpy = np.array(phrase)
- Read subgroups parallel bug?
- Poor performance of
H5Lget_name_by_idxorH5Literatewith large numbers of links - Running against BeeGFS
- File sizes of 1-2 GB
- Better strategy: Use HDF5 core VFD (
H5Pset_fapl_core) to load into memory and then do the iteration without I/O at memory speed.
- Poor performance of
- Help with broken file
- Sad story
- We talked about the HDF5 file state about a year ago
- There are currently no user-level transactions in HDF5, making file content vulnerable to application crash
- Familiarity with the file format specification and
h5debugcan help to inspect parts of an HDF5 file that is in an inconsistent state - A proper discovery tool of recoverable information would be great & we even
have ideas how to do it
There are comments in the file format specification such as this:
The ASCII character string "SNOD" is used to indicate the beginning of a symbol table node. This gives file consistency checking utilities a better chance of reconstructing a damaged file.
Alas, those "file consistency checking utilities" never materialized.
- Unable to open a Dataset in my HDF5 file
Tips, tricks, & insights
- How do HDF5-UDF work?
Currently, they are represented as chunked datasets with a single chunk. That's why they work fine with existing tools. The UDF itself is executed as part of the HDF5 filter pipeline. Its code is stored in the dataset blob data plus metadata and managed by the UDF handler.
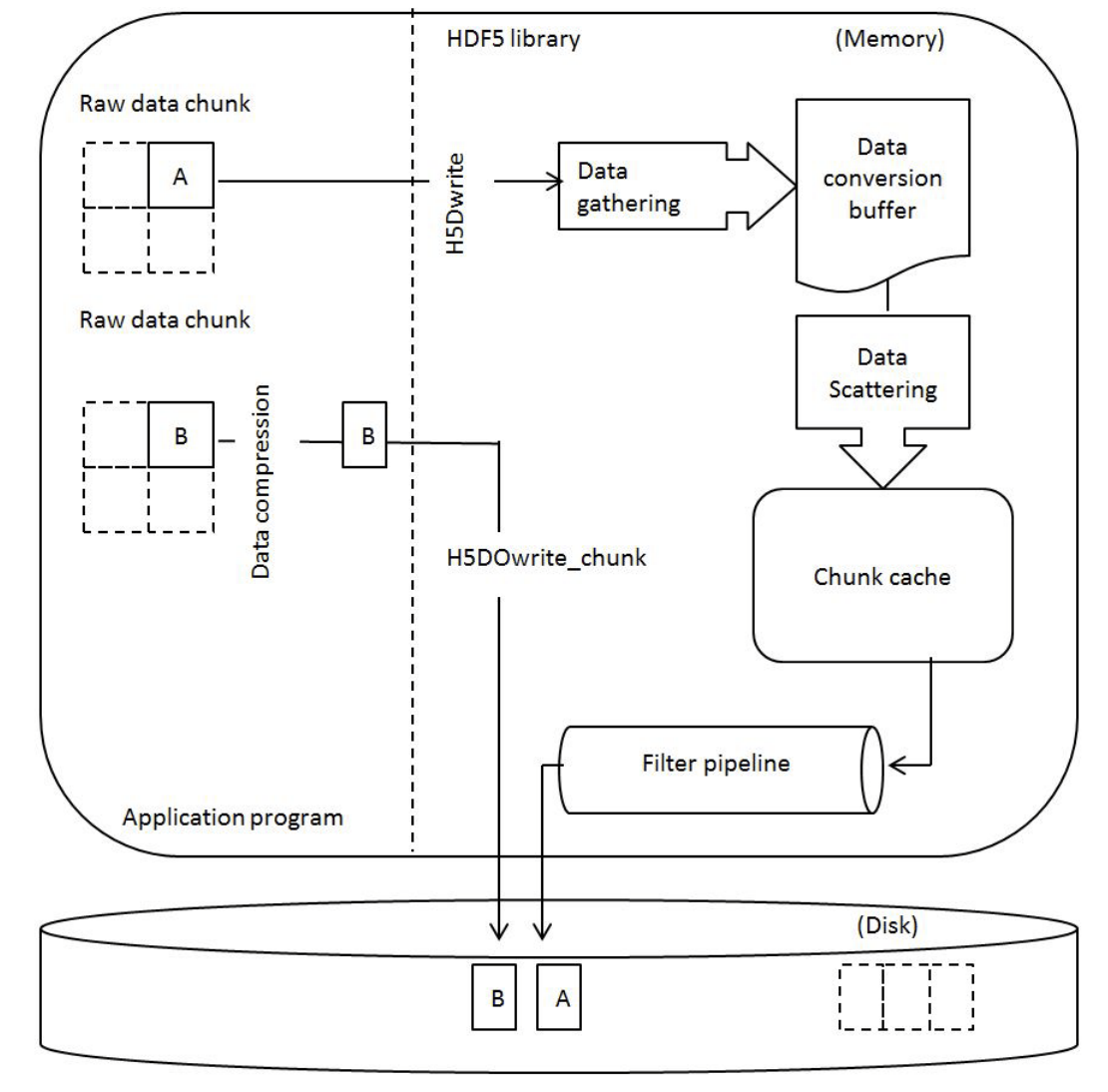
Figure 3:
H5Dwrite_chunk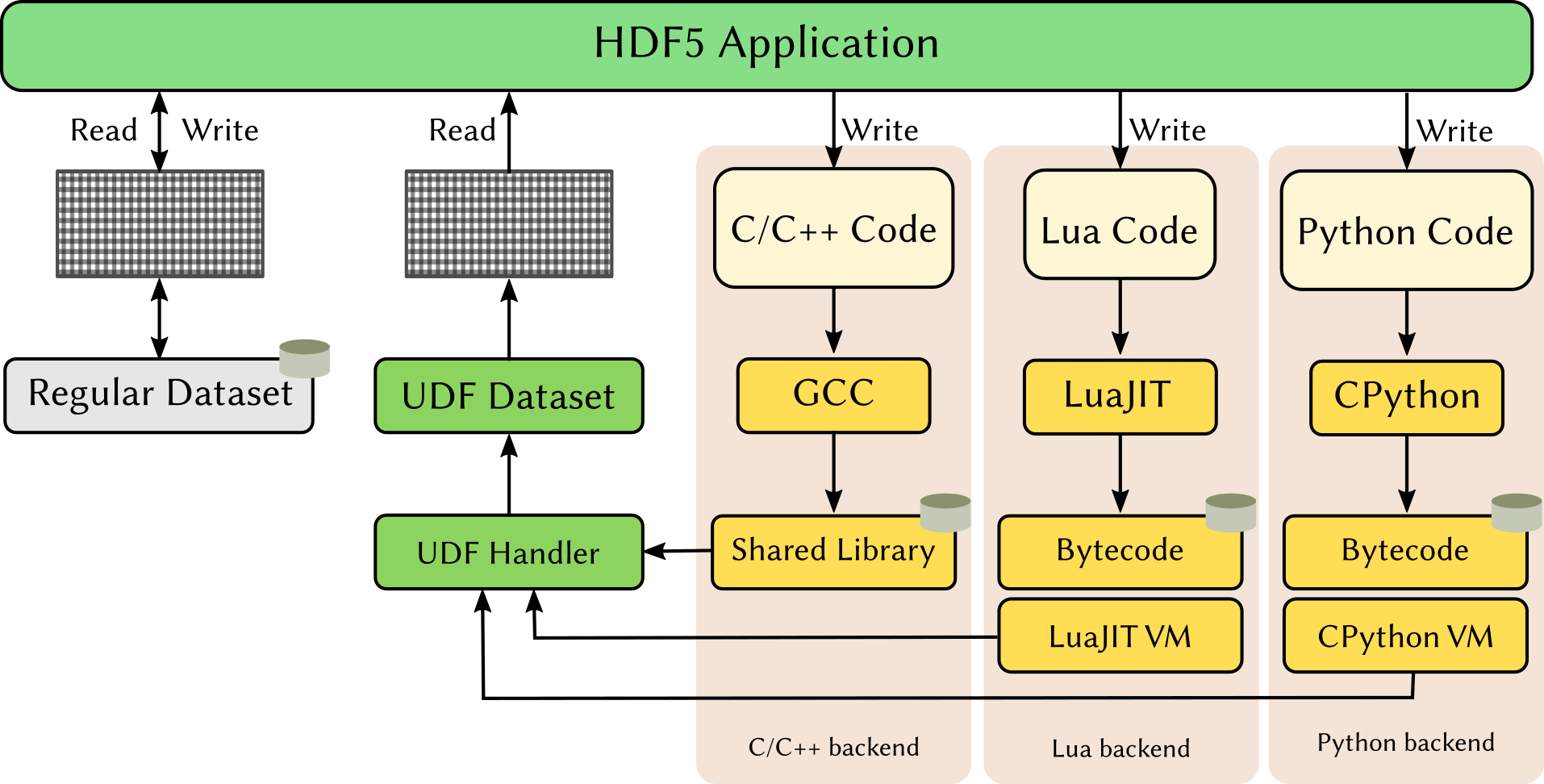
Figure 4: HDF5-UDF overview (Lucas C. Villa Real)
Example: Virtualization of CSV files through HDF5-UDF
def dynamic_dataset(): udf_data = lib.getData("GreatestAlbums") udf_dims = lib.getDims("GreatestAlbums") # The file is encoded as ISO-8859-1, so instruct Python about it with open("albumlist.csv", encoding="iso-8859-1") as f: # Read and ignore the header f.readline() for i, line in enumerate(f.readlines()): # Remove double-quotes and newlines around certain strings parts = [col.strip('"').strip("\n") for col in line.split(",")] udf_data[i].Number = int(parts[0]) udf_data[i].Year = int(parts[1]) lib.setString(udf_data[i].Album, parts[2].encode("utf-8")) lib.setString(udf_data[i].Artist, parts[3].encode("utf-8")) lib.setString(udf_data[i].Genre, parts[4].encode("utf-8")) lib.setString(udf_data[i].Subgenre, parts[5].encode("utf-8"))- Resources
Clinic 2022-02-15
Your Questions
- Q
- ???
Last week's highlights
- Announcements
- VOL tutorial postponed
- New date February 25th, 11:00 a.m. to 1:00 p.m. (Central)
- You can still register
- Check out this interesting blog post
- VOL tutorial postponed
- Forum
- Read compound data to buffer
- HDFView screenshot of a compound dataset
Simple & elegant HDFql solution
#include <iostream> #include "HDFql.hpp" struct data { unsigned long long timestamp; int order; int serial_number; double temperature; double pressure; int int_array[2][2]; }; int main(int argc, char *argv[]) { struct data values[4]; HDFql::variableRegister(values); HDFql::execute("select from h5ex_t_cmpd.h5 DS1 into memory 0"); for(int i = 0; i < 4; i++) { std::cout << "Timestamp=" << values[i].timestamp << std::endl; std::cout << "Order=" << values[i].order << std::endl; std::cout << "Serial number=" << values[i].serial_number << std::endl; std::cout << "Temperature=" << values[i].temperature << std::endl; std::cout << "Pressure=" << values[i].pressure << std::endl; std::cout << "IntArray[0][0]=" << values[i].int_array[0][0] << std::endl; std::cout << "IntArray[0][1]=" << values[i].int_array[0][1] << std::endl; std::cout << "IntArray[1][0]=" << values[i].int_array[1][0] << std::endl; std::cout << "IntArray[1][1]=" << values[i].int_array[1][1] << std::endl; } return 0; }
- Can we use external links to read-only data in writable files?
- Interesting follow-up from Thomas Kluyver (
h5pyproject)h5pydoes not currently exposeH5Pset_elink_acc_flags- Would be easy to do in the low-level API
- High-level API unclear
- Ambiguous error message; API failure due to insufficient permissions vs. non-existent file or directory
- Interesting follow-up from Thomas Kluyver (
- Read subgroups parallel bug?
- Goal: Determine the link names of blocks of subgroups in a group in parallel.
- Idea: Assign a link index range to each MPI process and fire away:
H5Lget_name_by_idx. - Issues:
- MPI hangs; not sure why…
H5Lget_name_by_idxis an expensive call, looping overidxmakes it worse- Parallel file systems don't like it
H5Literate+ callback would be better, but not much- To many small reads will be a drag on performance
- The real question is what the underlying use case is about.
- Maybe a dataset of object references instead of a group would be more suitable?
- Read compound data to buffer
Tips, tricks, & insights
- Do you need a link or a reference?
- HDF5 link
Explicit, unidirectional, named association between a source (HDF5 group) and a destination
name src -------> dst- HDF5 (object) reference
An HDF5 datatype whose values represent references (pointers) to HDF5 objects in the same or other HDF5 files.
&object
- Similarities
- Links can be used in the role of a (non-value) reference
- A link can be "de-referenced" by link traversal
- References can be used as implicit links
- A reference can be "traversed" by address/token resolution
- Links can be used in the role of a (non-value) reference
- Differences
- Links
- Are inseparable from groups and not values of an HDF5 datatype
- Have a name
- Can be dangling
- References
- Are values of an HDF5 datatype
- We can store them in attributes and datasets
- Have no name other than their implicit position
- Cannot dangle
- Links
Clinic 2022-02-08
Your Questions
- Q
- ???
- Q
Can a point selection be written to/read from a hypserslab selection? Does this work in parallel? Yes!
#include "hdf5.h" #include "mpi.h" #include <stdlib.h> int main(int argc, char** argv) { // boilerplate int retval = EXIT_SUCCESS; if (MPI_Init(&argc, &argv) != MPI_SUCCESS) { retval = EXIT_FAILURE; goto fail_mpi; } MPI_Comm comm = MPI_COMM_WORLD; int size, rank; if (MPI_Comm_size(comm, &size) != MPI_SUCCESS || MPI_Comm_rank(comm, &rank) != MPI_SUCCESS) { retval = EXIT_FAILURE; goto fail_file; } hid_t fapl = H5I_INVALID_HID; if ((fapl = H5Pcreate(H5P_FILE_ACCESS)) == H5I_INVALID_HID || H5Pset_fapl_mpio(fapl, comm, MPI_INFO_NULL) < 0) { retval = EXIT_FAILURE; goto fail_file; } hid_t file = H5I_INVALID_HID; if ((file = H5Fcreate("sel_par.h5", H5F_ACC_TRUNC, H5P_DEFAULT, fapl)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_file; } // 1D filespace, size of communicator hid_t fspace = H5I_INVALID_HID; if ((fspace = H5Screate_simple(1, (hsize_t[]) {2*size}, NULL)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_fspace; } hid_t dset = H5I_INVALID_HID; if ((dset = H5Dcreate(file, "ints", H5T_STD_I32LE, fspace, H5P_DEFAULT, H5P_DEFAULT, H5P_DEFAULT)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_dset; } // 2-element memory space hid_t mspace = H5I_INVALID_HID; if ((mspace = H5Screate_simple(1, (hsize_t[]) {2}, NULL)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_mspace; } int data[2] = { 2*rank, 2*rank+1 }; // 1. Make a (single) point selection in memory // 2. Make a hyperslab selection in the file // 3. Write if (H5Sselect_elements(mspace, H5S_SELECT_SET, 1, (hsize_t[]){0}) < 0 || H5Sselect_hyperslab(fspace, H5S_SELECT_SET, (hsize_t[]){2*rank}, NULL, (hsize_t[]){1}, (hsize_t[]){1}) < 0 || H5Dwrite(dset, H5T_NATIVE_INT, mspace, fspace, H5P_DEFAULT, data) < 0) { retval = EXIT_FAILURE; goto fail_write; } fail_write: H5Sclose(mspace); fail_mspace: H5Dclose(dset); fail_dset: H5Sclose(fspace); fail_fspace: H5Fclose(file); fail_file: if (fapl != H5I_INVALID_HID) H5Pclose(fapl); MPI_Barrier(comm); MPI_Finalize(); fail_mpi: return retval; }
Output:
penguin:~$ mpiexec -n 4 ./sel_par penguin:~$ h5dump sel_par.h5 HDF5 "sel_par.h5" { GROUP "/" { DATASET "ints" { DATATYPE H5T_STD_I32LE DATASPACE SIMPLE { ( 8 ) / ( 8 ) } DATA { (0): 0, 0, 2, 0, 4, 0, 6, 0 } } } }
Last week's highlights
- Announcements
- VOL tutorial postponed
- New date February 25th, 11:00 a.m. to 1:00 p.m. (Central)
- You can still register
- Check out this interesting blog post
- VOL tutorial postponed
- Forum
- Recover corrupted datasets
- Turns out that there was a single corrupted chunk
- Unclear how it got corrupted
- The user was able to recover the good chunks via
H5Dread_chunk
- Using GZIP compression does not compress, and cannot be opened by HDFView
- It appears that the user didn't appreciate the implications of using
H5Dwrite_chunk- Using this function tells the HDF5 library to "step aside" and let the user take full control
- Compression outside the library is fine, but you need to make sure that the right metadata is in place
- It appears that the user didn't appreciate the implications of using
- Why are there .err files next to test files in binary plugin download?
- Just artifacts for testing
- Could they be stripped from release binaries? Perhaps.
- Can we use external links to read-only data in writable files?
- Traversing external links typically involves opening another HDF5 file
- What's the access mode? (The link does NOT contain that information.)
- Default access mode: inherited from the link's "parent"
- Can be overwritten via a link access property list with
H5Pset_elink_acc_flags
- Compression filter that employes multiple datasets
- Use case: compression of unstructured mesh connectivity
- Involves multiple datasets
- The current filter extension API has no support for that
- Use case: compression of unstructured mesh connectivity
- Recover corrupted datasets
Tips, tricks, & insights
Back next time.
Clinic 2022-02-01
Your Questions
- Q
- ???
- Q
- Can a point selection be written to/read from a hypserslab selection?
Does this work in parallel?
- It appears to work for simple examples in sequential mode
- I have yet to try parallel mode
Last week's highlights
- Announcements
- Auth0 authentication issues resolved
- Let us know if you still have issues!
- VOL tutorial postponed
- New date February 25th, 11:00 a.m. to 1:00 p.m. (Central)
- You can still register
- Hermes 0.3.0-beta release
- Available on GitHub
- Most notable change: a publisher/subscriber API by Jamie Cernuda
- Stay tuned for an upcoming blog post
- Auth0 authentication issues resolved
- Forum
- ???
Tips, tricks, & insights
- HDFView binaries for Debian and Ubuntu
- They are well-hidden
:-(- It takes four clicks to get there (if you know where to go…)
- Current version (HDFView 3.1.3) link
- Download an unpack (
tar -zxvf ...) - Content Debian installer and
README.txt - Install w/
sudo dpkg -i hdfview_..._amd64deb - Installation directories
/opt/hdfview/[bin,lib,share] - Launch w/
/opt/hdfview/bin/HDFView(add to yourPATHas needed) - Does it need to be this hard?
- They are well-hidden
- One Stop HDF5 - HDF Lab
- Your registration w/ the HDF Group website is the ticket
- Home directory w/ 10 GB of free storage
- HSDS,
h5py,h5cc - Plenty of examples (Jupyter notebooks, Python scripts, etc.)
- Coming soon:
h5web
h5web
- Developed by ESRF
- React components for data visualization and exploration
- "HDFView for the browser"
- GitHub
- Supports HDF5 files in POSIX file systems & HSDS
- A JupyterHub plugin, jupyterlab-h5web, is available
Clinic 2022-01-25
Your Questions
- Q
- ???
- Q
- Can a point selection be written to/read from a hypserslab selection?
Does this work in parallel?
- It appears to work for simple examples in sequential mode
- I have yet to try parallel mode
Last week's highlights
- Announcements
- Auth0 authentication issues (apologies…)
- VOL tutorial postponed
- New date February 25th, 11:00 a.m. to 1:00 p.m. (Central)
- You can still register
- Auth0 authentication issues (apologies…)
- Forum
- Does the MPI driver lock the file?
- Yes, but I don't understand the user's description
I implemented a stride which tells the generation script how much work is being sent to each slave from the master. Now I have 4 datasets and each dataset has something like 400k entries. Each slave rank will write to all 4 datasets.
Now if I set the stride to a low value (10), the generation is way faster than if I set it to a big value (1024).
I wasn’t able to find how parallelism is exactly implemented. From the above behaviour it looks like the file is being locked which then blocks my whole program, especially if the stride is big (more time for the other ranks to run into a lock and be idle in between). Is that really the case? I write data continuously, so theoretically there is no need for a lock. Is is possible to tell the driver “don’t lock the file”?
- What's a 'stride'? (not a hyperslab stride…)
- Parallelism is implemented through MPI-I/O
- How does a file lock block the program?
- File locks can be disabled programmatically or via environment variable
- H5Datatype with variable length: How to set the values?
HDFql is adding support for variable-length datatypes in JAVA
// declare Java class that "mimics" the HDF5 compound dataset class Data { int myDimension; int myShapeType; int myInterpolationType; int myIntegrationType; int myNumberOfNormalComponents; int myNumberOfShearComponents; ArrayList myConnectivity; ArrayList myFaceConnectivity; } // declare variables Data write[] = new Data[1]; Data read[] = new Data[1]; // create HDF5 file 'myFile.h5' and use (i.e. open) it HDFql.execute("CREATE AND USE FILE myFile.h5"); // create compound dataset 'myDataset' HDFql.execute("CREATE DATASET myDataset AS COMPOUND(myDimension AS INT, myShapeType AS INT, myInterpolationType AS INT, myIntegrationType AS INT, myNumberOfNormalComponents AS INT, myNumberOfShearComponents AS INT, myConnectivity AS VARINT, myFaceConnectivity AS VARINT)"); // populate variable 'write' with dummy values write[0] = new Data(); write[0].myDimension = 1; write[0].myShapeType = 2; write[0].myInterpolationType = 3; write[0].myIntegrationType = 4; write[0].myNumberOfNormalComponents = 5; write[0].myNumberOfShearComponents = 6; write[0].myConnectivity = new ArrayList(); write[0].myConnectivity.add(10); write[0].myConnectivity.add(20); write[0].myFaceConnectivity = new ArrayList(); write[0].myFaceConnectivity.add(30); write[0].myFaceConnectivity.add(40); write[0].myFaceConnectivity.add(50); // write content of variable 'write' into dataset 'myDataset' HDFql.execute("INSERT INTO myDataset VALUES FROM MEMORY " + HDFql.variableRegister(write)); // read content of dataset 'myDataset' and populate variable 'read' with it HDFql.execute("SELECT FROM myDataset INTO MEMORY " + HDFql.variableRegister(read)); // display content of variable 'read' System.out.println("myDimension: " + read[0].myDimension); System.out.println("myShapeType: " + read[0].myShapeType); System.out.println("myInterpolationType: " + read[0].myInterpolationType); System.out.println("myIntegrationType: " + read[0].myIntegrationType); System.out.println("myNumberOfNormalComponents: " + read[0].myNumberOfNormalComponents); System.out.println("myNumberOfShearComponents: " + read[0].myNumberOfShearComponents); for(int i = 0; i < read[0].myConnectivity.size(); i++) { System.out.println("myConnectivity: " + read[0].myConnectivity.get(i)); } for(int i = 0; i < read[0].myFaceConnectivity.size(); i++) { System.out.println("myFaceConnectivity: " + read[0].myFaceConnectivity.get(i)); }
- Does the MPI driver lock the file?
Tips, tricks, & insights
- HDF5 - the better TXT format
- Source in GitHub repo
- Basic idea (Joe Lee): Store text files as compressed byte streams in HDF5 files.
- Pros:
- Passes the
difftest - Size reduction
- Portable metadata + rich annotation
- Unicode encoding
- Dataset region references
- Passes the
- Cons:
- Not suitable as editor back-ends
Clinic 2022-01-18
Your Questions
- Q
- ???
- Q
- Can a point selection be written to/read from a hypserslab selection?
Does this work in parallel?
- It appears to work for simple examples in sequential mode
- I have yet to try parallel mode
Last week's highlights
- Announcements
- VOL tutorial postponed
- New date February 25th, 11:00 a.m. to 1:00 p.m. (Central)
- You can still register
- VOL tutorial postponed
- Forum
- Is HSDS self-hosted?
- Can I host it myself?
- Yes, you can!
- References:
- Quick Start
Python:
h5pydviapip.import h5pyd as h5py ...
- Learn about the open source Highly Scalable Data Service (HSDS) Webinar
- HDF Lab
- Can I host it myself?
- convert seismic data (segy format) to HDF5 data?
- Not really a problem for "geeks"
- An Adaptable Seismic Data Format
- Seismic Data
- HDF5 doesn’t support writing VLENs with parallel I/O?
- Current (internal) VL representation is rather inefficient
- Fix/revise first, then parallelization
- Error when selecting and writing an element dataset hdf5 1.10.4
- (Bogus) Error message suggests that
H5T_NATIVE_DOUBLEis not a valid datatype - Maybe shared library not found or multiple copies?
- (Bogus) Error message suggests that
- Is HSDS self-hosted?
Tips, tricks, & insights
- Highly Scalable Data Service (HSDS)
- "HDF5 as a Service"
- REpresentational State Transfer (REST)
- HDF Lab has a few examples
- Let's do it! (from Emacs)
- HDF5 file "=" HSDS domain
Querying a domain
GET http://hsdshdflab.hdfgroup.org/?domain=/shared/tall.h5
{ "root": "g-d38053ea-3418fe27-5b08-db62bc-9076af", "class": "domain", "owner": "admin", "created": 1622930252.3698952, "limits": { "min_chunk_size": 1048576, "max_chunk_size": 4194304, "max_request_size": 104857600 }, "compressors": [ "blosclz", "lz4", "lz4hc", "gzip", "zstd", "deflate" ], "version": "0.7.0", "lastModified": 1623085764.3507726, "hrefs": [ { "rel": "self", "href": "http://hsdshdflab.hdfgroup.org/?domain=/shared/tall.h5" }, { "rel": "database", "href": "http://hsdshdflab.hdfgroup.org/datasets?domain=/shared/tall.h5" }, { "rel": "groupbase", "href": "http://hsdshdflab.hdfgroup.org/groups?domain=/shared/tall.h5" }, { "rel": "typebase", "href": "http://hsdshdflab.hdfgroup.org/datatypes?domain=/shared/tall.h5" }, { "rel": "root", "href": "http://hsdshdflab.hdfgroup.org/groups/g-d38053ea-3418fe27-5b08-db62bc-9076af?domain=/shared/tall.h5" }, { "rel": "acls", "href": "http://hsdshdflab.hdfgroup.org/acls?domain=/shared/tall.h5" }, { "rel": "parent", "href": "http://hsdshdflab.hdfgroup.org/?domain=hdflab2/shared" } ] } // GET http://hsdshdflab.hdfgroup.org/?domain=/shared/tall.h5 // HTTP/1.1 200 OK // Content-Type: application/json; charset=utf-8 // Date: Tue, 29 Nov 2022 18:05:26 GMT // Server: Highly Scalable Data Service (HSDS) for HDFLab // X-XSS-Protection: 1; mode=block // Content-Length: 1020 // Connection: keep-alive // Request duration: 1.576781sQuerying the HDF5 root group w/ resource ID
g-d38053ea-3418fe27-5b08-db62bc-9076afGET http://hsdshdflab.hdfgroup.org/groups/g-d38053ea-3418fe27-5b08-db62bc-9076af/links?domain=/shared/tall.h5
{ "links": [ { "class": "H5L_TYPE_HARD", "id": "g-d38053ea-3418fe27-3227-467313-8ebf63", "created": 1622930252.985488, "title": "g1", "collection": "groups", "target": "http://hsdshdflab.hdfgroup.org/groups/g-d38053ea-3418fe27-3227-467313-8ebf63?domain=/shared/tall.h5", "href": "http://hsdshdflab.hdfgroup.org/groups/g-d38053ea-3418fe27-5b08-db62bc-9076af/links/g1?domain=/shared/tall.h5" }, { "class": "H5L_TYPE_HARD", "id": "g-d38053ea-3418fe27-96ba-7678c2-3d4bcb", "created": 1622930252.5707703, "title": "g2", "collection": "groups", "target": "http://hsdshdflab.hdfgroup.org/groups/g-d38053ea-3418fe27-96ba-7678c2-3d4bcb?domain=/shared/tall.h5", "href": "http://hsdshdflab.hdfgroup.org/groups/g-d38053ea-3418fe27-5b08-db62bc-9076af/links/g2?domain=/shared/tall.h5" } ], "hrefs": [ { "rel": "self", "href": "http://hsdshdflab.hdfgroup.org/groups/g-d38053ea-3418fe27-5b08-db62bc-9076af/links?domain=/shared/tall.h5" }, { "rel": "home", "href": "http://hsdshdflab.hdfgroup.org/?domain=/shared/tall.h5" }, { "rel": "owner", "href": "http://hsdshdflab.hdfgroup.org/groups/g-d38053ea-3418fe27-5b08-db62bc-9076af?domain=/shared/tall.h5" } ] } // GET http://hsdshdflab.hdfgroup.org/groups/g-d38053ea-3418fe27-5b08-db62bc-9076af/links?domain=/shared/tall.h5 // HTTP/1.1 200 OK // Content-Type: application/json; charset=utf-8 // Date: Tue, 29 Nov 2022 18:05:26 GMT // Server: Highly Scalable Data Service (HSDS) for HDFLab // X-XSS-Protection: 1; mode=block // Content-Length: 1125 // Connection: keep-alive // Request duration: 0.149993sLet's look at a dataset
GET http://hsdshdflab.hdfgroup.org/datasets/d-d38053ea-3418fe27-cb7b-00379e-75d3e8?domain=/shared/tall.h5
{ "id": "d-d38053ea-3418fe27-cb7b-00379e-75d3e8", "root": "g-d38053ea-3418fe27-5b08-db62bc-9076af", "shape": { "class": "H5S_SIMPLE", "dims": [ 10 ], "maxdims": [ 10 ] }, "type": { "class": "H5T_FLOAT", "base": "H5T_IEEE_F32BE" }, "creationProperties": { "layout": { "class": "H5D_CHUNKED", "dims": [ 10 ] }, "fillTime": "H5D_FILL_TIME_ALLOC" }, "layout": { "class": "H5D_CHUNKED", "dims": [ 10 ] }, "attributeCount": 0, "created": 1622930252, "lastModified": 1622930252, "domain": "/shared/tall.h5", "hrefs": [ { "rel": "self", "href": "http://hsdshdflab.hdfgroup.org/datasets/d-d38053ea-3418fe27-cb7b-00379e-75d3e8?domain=/shared/tall.h5" }, { "rel": "root", "href": "http://hsdshdflab.hdfgroup.org/groups/g-d38053ea-3418fe27-5b08-db62bc-9076af?domain=/shared/tall.h5" }, { "rel": "home", "href": "http://hsdshdflab.hdfgroup.org/?domain=/shared/tall.h5" }, { "rel": "attributes", "href": "http://hsdshdflab.hdfgroup.org/datasets/d-d38053ea-3418fe27-cb7b-00379e-75d3e8/attributes?domain=/shared/tall.h5" }, { "rel": "data", "href": "http://hsdshdflab.hdfgroup.org/datasets/d-d38053ea-3418fe27-cb7b-00379e-75d3e8/value?domain=/shared/tall.h5" } ] } // GET http://hsdshdflab.hdfgroup.org/datasets/d-d38053ea-3418fe27-cb7b-00379e-75d3e8?domain=/shared/tall.h5 // HTTP/1.1 200 OK // Content-Type: application/json; charset=utf-8 // Date: Tue, 29 Nov 2022 18:05:26 GMT // Server: Highly Scalable Data Service (HSDS) for HDFLab // X-XSS-Protection: 1; mode=block // Content-Length: 1116 // Connection: keep-alive // Request duration: 0.215345s- Check it out with your favorite REST client!
Clinic 2022-01-11
Your Questions
- Q
- ???
- Q
- Can a point selection be written to/read from a hypserslab selection?
Does this work in parallel?
- It appears to work for simple examples in sequential mode
- I have yet to try parallel mode
Last week's highlights
- Announcements
- VOL tutorial postponed
- New date February 25th, 11:00 a.m. to 1:00 p.m. (Central)
- You can still register
- VOL tutorial postponed
- Forum
- select hyperslab of VL data
- Two issues:
- Getting the selections right
Dealing w/ VLEN data
struct s_data { uint64_t b; uint16_t a; }; struct ext_data3 { uint64_t a; uint32_t b; int16_t nelem; struct s_data data[3]; // <- ARRAY }; struct ext_data { uint64_t a; uint32_t b; int16_t nelem; struct s_data data[]; // <- VLEN };
- Nested compound (surface) datatype
- Attempted byte-stream representation as
\0-terminated VLEN string
- Two issues:
- Dynamically change the File Access Property List
- File access properties
- Vs. file creation properties
Set before file creation or file open
hid_t fapl = H5Pcreate(H5P_FILE_ACCESS); H5Pset_alignment(fapl, threshold, alignment); ... H5Fopen(..., fapl) or H5Fcreate(..., fapl) ...
- What is the use case for changing them dynamically?
- Wouldn't make sense for some properties, e.g., VFD
- Dynamic alignment changes, why?
- File access properties
- select hyperslab of VL data
Tips, tricks, & insights
- HDF5 snippets
- Developer productivity
- IntelliSense in VSCode
- Language Server Protocol (LSP)
- Emacs has support for LSP via
lsp-mode- Resource-intensive
- Not a templating mechanism
- YASnippet is a template system for Emacs
Easy to install and configure
(use-package yasnippet :custom (yas-triggers-in-field t) :config (setq yas-snippet-dirs "~/.emacs.d/snippets") (yas-global-mode 1))
- A (growing) set of snippets can be found here
- Demo
- Developer productivity
Clinic 2022-01-04
Your Questions
- Q
- ???
- Q
- Can a point selection be written to/read from a hypserslab selection?
Does this work in parallel?
- It appears to work for simple examples in sequential mode
- I have yet to try parallel mode
Last week's highlights
- Announcements
Happy New Year!
- Forum
- Repair corrupted file
- There's no general tool for that (yet)
Rigorous error checking and resource handling goes a long way
{ __label__ fail_file; hid_t file, group; char src_path[] = "/a/few/groups"; if ((file = H5Fcreate("o1.h5", H5F_ACC_TRUNC, H5P_DEFAULTx2)) == H5I_INVALID_HID) { ret_val = EXIT_FAILURE; goto fail_file; } // create a few groups { __label__ fail_group, fail_lcpl; hid_t lcpl; if ((lcpl = H5Pcreate(H5P_LINK_CREATE)) == H5I_INVALID_HID) { ret_val = EXIT_FAILURE; goto fail_lcpl; } if (H5Pset_create_intermediate_group(lcpl, 1) < 0) { ret_val = EXIT_FAILURE; goto fail_group; } if ((group = H5Gcreate(file, src_path, lcpl, H5P_DEFAULTx2)) == H5I_INVALID_HID) { ret_val = EXIT_FAILURE; goto fail_group; } H5Gclose(group); fail_group: H5Pclose(lcpl); fail_lcpl:; } // create a copy if (H5Ocopy(file, ".", file, "copy of", H5P_DEFAULTx2) < 0) { ret_val = EXIT_FAILURE; } H5Fclose(file); fail_file:; }- This looks pretty awkward, but there's some method to the madness…
- Repair corrupted file
Tips, tricks, & insights
- A GUI for HDFql
- HDFql is the La-Z-Boy of HDF5 interfaces
- SQL is convenient and concise because we say what we want (declarative) rather than how to do it (imperative).
Example (evaluate with
C-c C-c):CREATE TRUNCATE AND USE FILE my_file.h5 CREATE DATASET my_group/my_dataset AS double(3) ENABLE zlib LEVEL 0 VALUES(4, 8, 6) SELECT FROM DATASET my_group/my_dataset
Really?
h5dump -p my_file.h5
- Homework: What's the line count of an program written in C?
- Emacs supports the execution of source code blocks in Org mode
- HDFql comes with a command line interface
Combine the two w/ a snippet of Emacs Lisp code
;; We assume that HDFqlCLI is in the path and that libHDFql.so is in ;; the LD_LIBRARY_PATH. (defun org-babel-execute:hdfql (body params) "Execute a block of HDFql code with org-babel." (message "executing HDFql source code block") (org-babel-eval (format "HDFqlCLI --no-status --execute=\"%s\"" body) "")) (push '("hdfql" . sql) org-src-lang-modes) (add-to-list 'org-structure-template-alist '("hq" . "src hdfql"))
- The rest is cosmetics
- See this GitHub repo for HDF5 support in Emacs
- Fork and create a PR, if you are interested in pushing this forward!
- HDFql is the La-Z-Boy of HDF5 interfaces
Clinic 2021-12-21
Your Questions
- Q
- ???
- Q
- Can a point selection be written to/read from a hypserslab selection?
Does this work in parallel?
- It appears to work for simple examples in sequential mode
- I have yet to try parallel mode
Last week's highlights
- Announcements
Nothing to report.
- Forum
- Memory management in conversions of variable length data types
Reading data represented as HDF5 variable-length sequences. =>
hvl_ttypedef struct { size_t len; /**< Length of VL data (in base type units) */ void * p; /**< Pointer to VL data */ } hvl_t;
- Who owns the memory attached to
p? - The caller! Clean up w/
H5Dvlen_reclaim(pre-HDF5 1.12.x) orH5Treclaim(HDF5 1.12+)
- Read/write compound containing `std::string` using native C hdf5 lib
- Don't pass C++ objects as arguments to C library functions!
- You might get lucky, but you are relying on compiler peculiarities.
- Your luck will run out eventually.
- You might get lucky, but you are relying on compiler peculiarities.
typedef struct { int serial_no; std::string location; // CHANGED FROM char* to std::string double temperature; double pressure; } sensor_t;
- Works with
H5Dwritefor strings of a certain size, fails forH5Dread - See libc++'s implementation of
std::stringby Joel Laity for details.
- Don't pass C++ objects as arguments to C library functions!
- Merge 2 groups from the same h5 file
Simple example
? /G1/D + /G2/D ---> /G3/( Σ = /G1/D + G2/D )- In this simple example, we want to "append" the elements of the dataset
/G2/Dto the elements of the dataset/G1/D - Question: Is copying dataset elements problematic?
- YES
- Use virtual datasets! The also provides maximum flexibility in
defining Σ and mapping the constituent datasets.
- If you are using an older version of HDF5, you could define a dataset of region references to fake virtual datasets. This is much less convenient.
- NO
- Pedestrian approach: create a new (joint) dataset which can accommodate
the constituent datasets and read and write the elements from the
constituents.
- Wrinkle: The constituent datasets are too large and to fit into memory.
- Page your way through the constituents!
- Wrinkle: The constituent datasets are too large and to fit into memory.
- Memory management in conversions of variable length data types
Tips, tricks, & insights
- A GUI for HDFql
- HDFql is the La-Z-Boy of HDF5 interfaces
- SQL is convenient and concise because we say what we want (declarative) rather than how to do it (imperative).
Example (evaluate with
C-c C-c):CREATE TRUNCATE AND USE FILE my_file.h5 CREATE DATASET my_group/my_dataset AS double(3) ENABLE zlib LEVEL 0 VALUES(4, 8, 6) SELECT FROM DATASET my_group/my_dataset
Really?
h5dump -p my_file.h5
- Homework: What's the line count of an program written in C?
- Emacs supports the execution of source code blocks in Org mode
- HDFql comes with a command line interface
Combine the two w/ a snippet of Emacs Lisp code
;; We assume that HDFqlCLI is in the path and that libHDFql.so is in ;; the LD_LIBRARY_PATH. (defun org-babel-execute:hdfql (body params) "Execute a block of HDFql code with org-babel." (message "executing HDFql source code block") (org-babel-eval (format "HDFqlCLI --no-status --execute=\"%s\"" body) "")) (push '("hdfql" . sql) org-src-lang-modes) (add-to-list 'org-structure-template-alist '("hq" . "src hdfql"))
- The rest is cosmetics:
- Syntax highlighting ("font locking" in Emacs-speak)
- Auto-indentation
- Sessions
- Ping me (Gerd Heber), if you are interested in pushing this forward!
- HDFql is the La-Z-Boy of HDF5 interfaces
On behalf of The HDF Group, I wish you a Merry Christmas and a Happy New Year!
Stay safe & come back next year!
Clinic 2021-12-07
Your Questions
- Q
- ???
- Q
- Can a point selection be written to/read from a hypserslab selection?
Does this work in parallel?
- It appears to work for simple examples in sequential mode
- I have yet to try parallel mode
Last week's highlights
- Announcements
- We had a great webinar Accelerate I/O operations with Hermes
- Stay tuned for the recording on YouTube
- The Hermes project now has its forum category
- Follow announcements, ask questions, participate!
- Release of HDF5-1.13.0
- An odd release number?
- Experimental vs. maintenance releases see here
- "Experimental" is not a fig leaf for "shoddy"
- Experimental releases receive as much TLC as maintenance releases
- Highlights:
- VOL layer updates (DAOS, pass-through, async.)
- VFD layer updates
- Dynamic loading
- GPUDirect VFD
- Performance improvements
h5dwalktool[ bin]$ mpiexec -n 4 ./h5dwalk -o show-h5dump-h5files.log -T ./h5dump $HOME/Sandbox/HDF5/GITHUB/hdf5/tools/testfiles [ bin]$ more show-h5dump-h5files.log --------- Command: ./h5dump -n /home/riwarren/Sandbox/HDF5/GITHUB/hdf5/tools/testfiles/tnestedcmpddt.h5 HDF5 "/home/riwarren/Sandbox/HDF5/GITHUB/hdf5/tools/testfiles/tnestedcmpddt.h5" { FILE_CONTENTS { group / dataset /dset1 dataset /dset2 dataset /dset4 dataset /dset5 datatype /enumtype group /group1 dataset /group1/dset3 datatype /type1 } } ...
- An odd release number?
- VOL tutorial moved to January 14, 2022!
- Covers the basics needed to construct a simple terminal VOL connector
- Great New Year's resolution
;-)
- We had a great webinar Accelerate I/O operations with Hermes
- Forum
- Working with packed 12-bit integers
- Two basic approaches: packing or filtering
- H5TurboPFor
- HDF5 frontend for TurboPFor: Fastest Integer Compression
- H5Datatype with variable length: How to set the values?
- Too many half-baked HDF5 Java interfaces (including our own)
- How can we better engage with that community?
- HDFql?
- Which layout shall I use?
- Acquiring a lot of small (< 8K) messages
- Which (dataset) layout is best for performance?
- What is layout?
- It depends…
- How is performance measured?
- How will the messages be accessed?
- Controlling BTree parameters for performance reasons
- Import large number of images (~5 million) as chunked datasets
- ~10-20 million groups for indexing
- Can B-tree parameters do magic? (No)
- Good intro to B-trees
Two kinds of B-trees, file-wide configuration via FCPL
// group links herr_t H5Pset_sym_k(hid_t plist_id, unsigned ik, unsigned lk); // dataset chunk index herr_t herr_t H5Pset_istore_k(hid_t plist_id, unsigned ik);
- Other potential remedies
- File format improvements
- Reduce the number of objects by stacking images, e.g., by resolution
- VFD SWMR beta 1 release
- Will the HDF5 SWMR VFD be a plugin?
I don't know for sure.- No. See Dana's response.
- Will the HDF5 SWMR VFD be a plugin?
- Virtual Data Set
For our application, we need to return an error in case the caller tries to read data from a VDS and some of the referenced files that store the requested data are not available.
- Currently, users cannot change the error behavior of VDS functions
- Pedestrian approach: parse the VDS metadata to detect missing files
- Working with packed 12-bit integers
Tips, tricks, & insights
No time for that, today.
Clinic 2021-11-23
Your Questions
- Q
- ???
- Q
Under Compatibility and Performance Issues we say
Not all HDF5-1.10 releases are compatible.
What does that mean and why?
- API incompatibility (not file format!) introduced in HDF5 1.10.3
- Q
- Can a point selection be written to/read from a hypserslab selection?
Does this work in parallel?
- It appears to work for simple examples in sequential mode:
#include "hdf5.h" #include <stdlib.h> int main() { __label__ fail_file, fail_fspace, fail_dset; int retval = EXIT_SUCCESS; hid_t file, fspace, dset, mspace; int data[] = {0, 1, 2, 3, 4, 5, 6, 7, 8, 9}; if((file = H5Fcreate("sel.h5", H5F_ACC_TRUNC, H5P_DEFAULT, H5P_DEFAULT)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_file; } if ((fspace = H5Screate_simple(1, (hsize_t[]) {10}, NULL)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_fspace; } if ((dset = H5Dcreate(file, "ints", H5T_STD_I32LE, fspace, H5P_DEFAULT, H5P_DEFAULT, H5P_DEFAULT)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_dset; } if ((mspace = H5Scopy(fspace)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_copy; } // 1. Make a point selection in memory // 2. Make a hyperslab selection in the file // 3. Write if (H5Sselect_elements(mspace, H5S_SELECT_SET, 3, (hsize_t[]){3, 1, 6}) < 0 || H5Sselect_hyperslab(fspace, H5S_SELECT_SET, (hsize_t[]){4}, NULL, (hsize_t[]){1}, (hsize_t[]){3}) < 0 || H5Dwrite(dset, H5T_NATIVE_INT, mspace, fspace, H5P_DEFAULT, data) < 0) { retval = EXIT_FAILURE; goto fail_write; } fail_write: H5Sclose(mspace); fail_copy: H5Dclose(dset); fail_dset: H5Sclose(fspace); fail_fspace: H5Fclose(file); fail_file: return retval; }
- The output file produce looks like this:
HDF5 "sel.h5" {
GROUP "/" {
DATASET "ints" {
DATATYPE H5T_STD_I32LE
DATASPACE SIMPLE { ( 10 ) / ( 10 ) }
DATA {
(0): 0, 0, 0, 0, 3, 1, 6, 0, 0, 0
}
}
}
}
- I have yet to try parallel mode
Last week's highlights
- Announcements
- Accelerate I/O operations with Hermes
- Hermes 0.1.0-beta
- Distributed I/O buffering for storage hierarchies
- GitHub & Getting Started Guide
- Register for the webinar on December 1, 2021 11:00 AM (Central)!
- Try the HDF5 SWMR VFD Beta!
- Accelerate I/O operations with Hermes
- Forum
- Reference Manual in Doxygen
- We've been experimenting with full-text search
- Not a replacement for the Doxygen-native search
- What do you think?
- Working with packed 12-bit integers
- Two basic approaches: packing or filtering
- H5TurboPFor
- HDF5 frontend for TurboPFor: Fastest Integer Compression
- H5Datatype with variable length: How to set the values?
- Too many half-baked HDF5 Java interfaces (including our own)
- How can we better engage with that community?
- HDFql?
- Which layout shall I use?
- Acquiring a lot of small (< 8K) messages
- Which (dataset) layout is best for performance?
- It depends…
- How is performance measured?
- How will the messages be accessed?
- Controlling BTree parameters for performance reasons
- Import large number of images (~5 million) as chunked datasets
- ~10-20 million groups for indexing
- Can B-tree parameters do magic? (No)
- Potential remedies
- File format improvements
- Reduce the number of objects by stacking images, e.g., by resolution
- VFD SWMR beta 1 release
- Will the HDF5 SWMR VFD be a plugin?
- I don't know for sure.
- Will the HDF5 SWMR VFD be a plugin?
- Reference Manual in Doxygen
Tips, tricks, & insights
No time for that, today.
Clinic 2021-11-16
Your Questions
- Q
- ???
- Q
Under Compatibility and Performance Issues we say
Not all HDF5-1.10 releases are compatible.
What does that mean and why?
- Q
- Can a point selection be written to/read from a hypserslab selection? Does this work in parallel?
Last week's highlights
- Announcements
- Accelerate I/O operations with Hermes
- Hermes 0.1.0-beta
- Distributed I/O buffering for storage hierarchies
- GitHub & Getting Started Guide
- Register for the webinar on December 1, 2021 11:00 AM (Central)!
- Try the HDF5 SWMR VFD Beta!
- Accelerate I/O operations with Hermes
- Forum
- VFD SWMR beta 1 release
- Part of HDF5 1.13?
- No and yes: de-coupling of VFDs and library releases
- HDF5FunctionArgumentException: Inappropriate type for Datatype.CLASSARRAY
- Drink responsibly & don't go crazy with those datatypes!
- Dynamically Loaded Filters on Mac OS
- The joy of being different
- H5Datatype with variable length: How to set the values?
- The joy and frustration of language bindings to the HDF5 C-API
- Open HDF5 when it is already opened in HDFVIEW
- Solution:
export HDF5_USE_FILE_LOCKING="FALSE"
- VFD SWMR beta 1 release
Tips, tricks, & insights
- Mochi - 2021 R&D100 Winner
- Mochi project page
- Collaboration between ANL, LANL, CMU, and The HDF Group
- See Jerome Soumagne's HUG 2021 presentation
- Changes in scientific workflows
- Composable data services and building blocks
- Micros-services rather than monoliths
- A refined toolset for modern architectures and demanding applications
- Who wants to share their favorite hack/trick?
Clinic 2021-11-09
Your Questions
- Q
- ???
- Q
Under Compatibility and Performance Issues we say
Not all HDF5-1.10 releases are compatible.
What does that mean and why?
- Q
- Can a point selection be written to/read from a hypserslab selection? Does this work in parallel?
Last week's highlights
- Announcements
- Accelerate I/O operations with Hermes
- Hermes 0.1.0-beta
- Distributed I/O buffering for storage hierarchies
- GitHub & Getting Started Guide
- Stay tuned for a webinar on December 1, 2021
- Try the HDF5 SWMR VFD Beta!
- Accelerate I/O operations with Hermes
- Forum
- Crash when writing parallel compressed chunks
- Jordan committed a fix for an old
MPI_ERR_TRUNCATEissue/crash - Merging to development branches of HDF5 1.13, 1.12, 1.10
- If you've been affected by this, give it a try!
- Jordan committed a fix for an old
- Sorting of members of a compound type
- User has to write elements of a compound datatype one at a time
- "Partial I/O gets in the way" - sorting fields by name or offset
- Happens on each write call => overhead
- Can this be avoided? User might provide a patch…
- Read/write specific coordinates in multi-dimensional dataset?
Thomas is looking for use cases from
h5pyusersarr = ds.points[[(0, 0), (1, 0), (2, 2)]] # Read ds.points[[(0, 0), (1, 0), (2, 2)]] = [1, 2, 3] # Write
- See his PR
- Reading variable length data from hdf5 file C++ API
- The "curse" of variable-length sequence data = the loss of memory contiguity
- C++20
std::spanseems to be the way to go, but how many codes are C++20? - Important: The cleanup of buffers into which VLEN data is read is the
caller's responsibility! Use
H5Dvlen_reclaim(HDF5 1.10-) orH5Treclaim(HDF5 1.12+). - Got
milkmatchingH5DreadandH5Dwrite?
- Crash when writing parallel compressed chunks
Tips, tricks, & insights
We didn't get to this last time…
H5Dread/H5DwriteSymmetry
Syntax
herr_t H5Dwrite ( hid_t dset_id, hid_t mem_type_id, // the library "knows" the in-file datatype hid_t mem_space_id, hid_t file_space_id, hid_t dxpl_id, const void* buf ); herr_t H5Dread ( hid_t dset_id, hid_t mem_type_id, // the library "knows" the in-file datatype hid_t mem_space_id, hid_t file_space_id, hid_t dxpl_id, void* buf );
- Necessary conditions for this to work out
- The in-memory (element) datatype must be convertible to/from the in-file
datatype. (With the exception of VLEN strings, VLEN types a la
hvl_tare not convertible to ragged arrays!) - The dataspace selections in-memory and in the file must have the same
number of selected elements. (Be careful when using
H5S_ALLfor one ofmem_space_idorfile_space_id!) - The buffer must be big enough to hold at least the number of selected
elements (in their native representation).
- For parallel, the number of elements written/read by this MPI rank
- The in-memory (element) datatype must be convertible to/from the in-file
datatype. (With the exception of VLEN strings, VLEN types a la
Clinic 2021-11-02
Your Questions
- Q
- ???
- Q
Under Compatibility and Performance Issues we say
Not all HDF5-1.10 releases are compatible.
What does that mean and why?
- Q
- Can a point selection be written to/read from a hypserslab selection? Does this work in parallel?
Last week's highlights
- Announcements
- HDF5 1.10.8 Release
- Release notes
- CMake no longer builds the C++ library by default
- HDF5 now requires Visual Studio 2015 or greater
- On macOS, Universal Binaries can now be built
- CMake option to build the HDF filter plugins project as an external project
- Autotools and CMake target added to produce doxygen generated documentation
- CMake option to statically link gcc libs with MinGW
- File locking now works on Windows
- Improved performance of
H5Sget_select_elem_pointlist - Detection of simple data transform function
"x"
- Interesting figure
- Under Compatibility and Performance Issues is
- Release notes
- Try the HDF5 SWMR VFD Beta!
- HDF5 1.10.8 Release
- Forum
H5Dget_chunk_infoperformance for many chunks?
- Task: Get all of the chunk file offsets + sizes
- Solution:
H5Dchunk_iter - Caveat: Currently only available in the
developmentbranch - Note: We covered this function and an example in our clinic on
- Open HDF5 when it is already opened in HDFVIEW
Is there a way (probably file access property) to open the file multiple times (especially when it is opened in HdfView) and allow to read/write it? May the problem be solved if I build hdf5 with multithreads option ON ?
- Except for specific use cases (SWMR), this is a bad idea
- Why? Remember this figure?

- Append HDF5 files in parallel
I have thousands of HDF5 files that need to be merged into a single file. Merging is simply to append all groups and datasets of one file after another in a new output file. The group names of the input files are all different from one another. In addition, all datasets are chunked and compressed.
My question is how do I merge the files in parallel?
My implementation consists of the following steps: …
- That's a tough one
- Two options
- Don't copy any data, just reference existing data (via external links)
- Copy data as fast as you can
- (MPI) parallel make this more complicated
- Reading variable length data from hdf5 file C++ API
- Got
milkmatchingH5DreadandH5Dwrite?
- Got
Tips, tricks, & insights
H5Dread/H5DwriteSymmetry
Syntax
herr_t H5Dwrite ( hid_t dset_id, hid_t mem_type_id, hid_t mem_space_id, hid_t file_space_id, hid_t dxpl_id, const void* buf ); herr_t H5Dread ( hid_t dset_id, hid_t mem_type_id, hid_t mem_space_id, hid_t file_space_id, hid_t dxpl_id, void* buf );
- Necessary conditions for this to work out
- The in-memory (element) datatype must be convertible to/from the in-file datatype.
- The dataspace selections in-memory and in the file must have the same number of selected elements.
- The buffer must be big enough to hold at least the number of selected
elements (in their native representation).
- For parallel, the number of elements written/read by this MPI rank
Clinic 2021-10-28
Your Questions
- Q
- ???
Last week's highlights
- Announcements
- HUG 2021 Videos are showing up on YouTube
- Submit comments/questions/criticisms to the organizing committee
- HUG 2021 Videos are showing up on YouTube
- Forum
- HDFView 3.1.0 error displaying dataset with np.float16 data
- HDF5 has supported user-defined floating-point types from day one
- Good example of poor execution on our part!
- GPUs have rekindled the interest in reduced floating-point representations
- Announcing development of the Enterprise Support Edition of HDF5
- Good example of an own goal / publicity disaster
- One more time: There is no Enterprise Support Edition of HDF5!
- However, support and consulting agreements are available from The HDF Group for enterprises and other organizations
- There is only one development line of HDF5 (data model, file format, library)
& it is available on GitHub
- Hence, it is nonsensical to speak of 'HDF5 editions'
- End of story
- HDFView 3.1.0 error displaying dataset with np.float16 data
Tips, tricks, & insights
- Who is afraid of
h5debug?
- A useful tool to explore the "guts" of the HDF5 file format
- There's even a nice guided tour by Quincey Koziol from 2003
- HDF5 1.4.5 was released
- HDF5 1.6.0 was released
Compiling and running
example1.cproduces this output:%h5debug example1.h5 Reading signature at address 0 (rel) File Super Block... File name (as opened): example1.h5 File name (after resolving symlinks): example1.h5 File access flags 0x00000000 File open reference count: 1 Address of super block: 0 (abs) Size of userblock: 0 bytes Superblock version number: 0 Free list version number: 0 Root group symbol table entry version number: 0 Shared header version number: 0 Size of file offsets (haddr_t type): 8 bytes Size of file lengths (hsize_t type): 8 bytes Symbol table leaf node 1/2 rank: 4 Symbol table internal node 1/2 rank: 16 Indexed storage internal node 1/2 rank: 32 File status flags: 0x00 Superblock extension address: 18446744073709551615 (rel) Shared object header message table address: 18446744073709551615 (rel) Shared object header message version number: 0 Number of shared object header message indexes: 0 Address of driver information block: 18446744073709551615 (rel) Root group symbol table entry: Name offset into private heap: 0 Object header address: 96 Cache info type: Symbol Table Cached entry information: B-tree address: 136 Heap address: 680- It matches the output from 2003 except for
- The root group's object header address is 96 (in 2021) vs. 928 (in 2003)
- The root's B-tree is at 136 vs. 384
- The root group's local heap is at 680 vs. 96
- Happy HDF5 exploring!
Clinic 2021-10-19
Your Questions
- Q
- ???
Last week's highlights
- Announcements
- Forum
- Dataset Insert Vector of Compound Type
Memory-contiguous vs. non-contiguous layout
struct Instruction { float timestamp; Key keys[MAX_KEYS]; // -> H5T_ARRAY }; struct Instruction { float timestamp; Key* keys; // -> H5T_VLEN };
- Memory mapping / Paging via HDF5 & VFD?
hid_t H5Dread_mapped(..., void** buf);- Returns a pointer to a memory-mapped region
- If the region is writable, changes will be propagated to the file (instead of
calling
H5Dwrite)
- External links & VFDs
- What if an external link requires a different VFD for its resolution?
- Current approach: set it manually via
H5Pset_elink_fapl()after figuring out that that's required. Yikes! - Add VFD-plugins and this gets out of hand quickly…
- Why increasing
rdcc_nbytesandrdcc_nslotswill result in a decrease in indexing performance?
- See RFC: Setting Raw Data Chunk Cache Parameters in HDF5 for guidance
- Potential
h5pyissue
- Fletcher32 filter on variable length string datasets (not suitable for filters)
- Triggered by change introduced in 1.12.1
- Caution: Filters and variable-length data
- Dataset Insert Vector of Compound Type
Tips, tricks, & insights
- Something's compressed:
#include "hdf5.h" #include <stdio.h> #include <stdlib.h> int main() { __label__ fail_file, fail_dtype, fail_dspace, fail_dcpl, fail_dset, fail_write; int retval = EXIT_SUCCESS; hid_t file, dspace, dtype, dcpl, dset; if ((file = H5Fcreate("vlen.h5", H5F_ACC_TRUNC, H5P_DEFAULT, H5P_DEFAULT)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_file; } if ((dtype = H5Tvlen_create(H5T_STD_I32LE)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_dtype; } if ((dspace = H5Screate_simple(1, (hsize_t[]){2048}, (hsize_t[]){H5S_UNLIMITED})) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_dspace; } if ((dcpl = H5Pcreate(H5P_DATASET_CREATE)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_dcpl; } if (H5Pset_chunk(dcpl, 1, (hsize_t[]) {1024}) < 0 || H5Pset_deflate(dcpl, 1) < 0 //H5Pset_fletcher32(dcpl) < 0 ) { retval = EXIT_FAILURE; goto fail_dset; } if ((dset = H5Dcreate(file, "dset", dtype, dspace, H5P_DEFAULT, dcpl, H5P_DEFAULT)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_dset; } { int data[] = {0, 1, 2, 3, 4, 5, 6, 7, 8, 9}; size_t offset[] = {0, 1, 3, 6}; hvl_t buf[2048]; size_t i; // create an array that looks like this: // { {0}, {1,2}, {3,4,5}, {6,7,8,9}, ...} for (i = 0; i < 2048; ++i) { size_t rem = i%4; buf[i].len = 1 + rem; buf[i].p = data + offset[rem]; } if (H5Dwrite(dset, dtype, H5S_ALL, H5S_ALL, H5P_DEFAULT, buf) < 0) { retval = EXIT_FAILURE; goto fail_write; } } fail_write: H5Dclose(dset); fail_dset: H5Pclose(dcpl); fail_dcpl: H5Sclose(dspace); fail_dspace: H5Tclose(dtype); fail_dtype: H5Fclose(file); fail_file: return retval; }
h5dump -pBH vlen.h5
HDF5 "vlen.h5" {
SUPER_BLOCK {
SUPERBLOCK_VERSION 0
FREELIST_VERSION 0
SYMBOLTABLE_VERSION 0
OBJECTHEADER_VERSION 0
OFFSET_SIZE 8
LENGTH_SIZE 8
BTREE_RANK 16
BTREE_LEAF 4
ISTORE_K 32
FILE_SPACE_STRATEGY H5F_FSPACE_STRATEGY_FSM_AGGR
FREE_SPACE_PERSIST FALSE
FREE_SPACE_SECTION_THRESHOLD 1
FILE_SPACE_PAGE_SIZE 4096
USER_BLOCK {
USERBLOCK_SIZE 0
}
}
GROUP "/" {
DATASET "dset" {
DATATYPE H5T_VLEN { H5T_STD_I32LE}
DATASPACE SIMPLE { ( 2048 ) / ( H5S_UNLIMITED ) }
STORAGE_LAYOUT {
CHUNKED ( 1024 )
SIZE 5772 (5.677:1 COMPRESSION)
}
FILTERS {
COMPRESSION DEFLATE { LEVEL 1 }
}
FILLVALUE {
FILL_TIME H5D_FILL_TIME_ALLOC
VALUE H5D_FILL_VALUE_DEFAULT
}
ALLOCATION_TIME {
H5D_ALLOC_TIME_INCR
}
}
}
}
- N.B. What's compressed are the in-file counterparts of
hvl_tstructures, not the integer sequences! - Filtering fails, if we enable Fletcher32
Clinic 2021-09-28
Your Questions
- Q
- Will the HDF5 1.12.1 file locking changes be brought to 1.10.8?
- Did Elena will answer that?
Last week's highlights
- Announcements
- HUG 2021 Agenda posted
- October 12-15, 2021, 9:00 AM - 1:30 PM (CDT)
- Webinar Announcement: New Features in the HDF5 1.13.0 Release
- HUG 2021 Agenda posted
- Forum
- Possible to change email address here?
- Lori?
- Retrieve property list from object
- Property lists come in several flavors (access, creation, transfer)
- Only creation property lists are store in the file
- Dynamic Plugins require application to link to shared library
- GitHub issue
- Plugin loading happens via
dlopen - Calling the HDF5 library from a plugin
- Why would applications do that? Element type, chunk size, …
- API shortcomings?
- Object timestamps - useful or not?
- Only a minority seems to care
- Limited implementation
- Bit-for-bit reproducibility
- Clear communication before changing defaults is crucial
- Only a minority seems to care
- Possible to change email address here?
Tips, tricks, & insights
- HDF5 references
- HDF5 datatype
- Pre-HDF5 1.12.0 referents limited to dataset regions and objects
- Starting w/ HDF5 1.12.0 referents can be HDF5 attributes
- Support for querying and indexing
- API clean-up
- Basic life cycle examples in RM
Clinic 2021-09-21
- Elena's slides on HDF5 Chunking and Compression - Performance issues
Clinic 2021-08-31
Your Questions
- Q
- Will the HDF5 1.12.1 file locking changes be brought to 1.10.8?
- Elena will answer that next week!
Tips, tricks, & insights
- Using a custom filter
#include "hdf5.h" #include <stdio.h> #include <stdlib.h> // an identity filter function which just prints "helpful" messages size_t filter(unsigned int flags, size_t cd_nelmts, const unsigned int cd_values[], size_t nbytes, size_t *buf_size, void **buf) { buf_size = 0; if (flags & H5Z_FLAG_REVERSE) { // read data, e.g., decompress data // ... printf("Decompressing...\n"); } else { // write data, e.g., compress data // ... printf("Compressing...\n"); } return nbytes; } int main() { // boilerplate __label__ fail_register, fail_file, fail_dspace, fail_dcpl, fail_dset, fail_write; int retval = EXIT_SUCCESS; hid_t file, dspace, dcpl, dset; // custom filter H5Z_class_t cls; cls.version = H5Z_CLASS_T_VERS; cls.id = 256; cls.encoder_present = 1; cls.decoder_present = 1; cls.name = "Identity filter"; cls.can_apply = NULL; cls.set_local = NULL; cls.filter = &filter; // register the filter if (H5Zregister(&cls) < 0) { retval = EXIT_FAILURE; goto fail_register; } if ((file = H5Fcreate("filter.h5", H5F_ACC_TRUNC, H5P_DEFAULT, H5P_DEFAULT)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_file; } if ((dspace = H5Screate_simple(1, (hsize_t[]){2048}, (hsize_t[]){H5S_UNLIMITED})) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_dspace; } if ((dcpl = H5Pcreate(H5P_DATASET_CREATE)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_dcpl; } // play with early chunk allocation and fill time if (H5Pset_filter(dcpl, cls.id, 0|H5Z_FLAG_MANDATORY, 0, NULL) < 0 || //H5Pset_alloc_time(dcpl, H5D_ALLOC_TIME_EARLY) < 0 || //H5Pset_fill_time(dcpl, H5D_FILL_TIME_NEVER) < 0 || H5Pset_chunk(dcpl, 1, (hsize_t[]) {1024}) < 0) { retval = EXIT_FAILURE; goto fail_dset; } if ((dset = H5Dcreate(file, "dset", H5T_STD_I32LE, dspace, H5P_DEFAULT, dcpl, H5P_DEFAULT)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_dset; } // write something to trigger the "compression" of two chunks { int data[2048]; if (H5Dwrite(dset, H5T_NATIVE_INT, H5S_ALL, H5S_ALL, H5P_DEFAULT, data) < 0) { retval = EXIT_FAILURE; goto fail_write; } } // housekeeping fail_write: H5Dclose(dset); fail_dset: H5Pclose(dcpl); fail_dcpl: H5Sclose(dspace); fail_dspace: H5Fclose(file); fail_file: // unregister the filter if (H5Zunregister(cls.id) < 0) { retval = EXIT_FAILURE; } fail_register: return retval; }
Clinic 2021-08-24
Your Questions
- Q
- Will the HDF5 1.12.1 file locking changes be brought to 1.10.8?
Last week's highlights
- Announcements
- I didn't see any
- Forum
- Append to compound dataset
- It is tempting to think about 1D compound datasets as tables
- Reference Manual in Doxygen
- RM H5Z
- Don't let Word & co. pollute your documentation!
- It's not someone else being evil, it's us acting like fools!
- Append to compound dataset
Tips, tricks, & insights
- HDF5 Compound Datasets and (Relational) Tables: Don't be fooled!
- Append to compound dataset
- 'Row' as in 'table row' or 'matrix row' share the same spelling, but that's
where the similarity ends!
- HDF5 datasets are not tables
#include "hdf5.h" #include <stdlib.h> int main() { __label__ fail_file, fail_dspace, fail_dset, fail_extent; int retval = EXIT_SUCCESS; hid_t file, dspace, dcpl, dset; if ((file = H5Fcreate("foo.h5", H5F_ACC_TRUNC, H5P_DEFAULT, H5P_DEFAULT)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_file; } // create a 1D dataspace of indefinite extent, initial extent 0 (elements) if ((dspace = H5Screate_simple(1, (hsize_t[]){0}, (hsize_t[]){H5S_UNLIMITED})) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_dspace; } // allocate space in the file in batches of 1024 dataset elements if ((dcpl = H5Pcreate(H5P_DATASET_CREATE)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_dcpl; } if (H5Pset_chunk(dcpl, 1, (hsize_t[]){1024}) < 0) { retval = EXIT_FAILURE; goto fail_dset; } // create the dataset // (replace H5T_STD_I32LE with your favorite datatype) if ((dset = H5Dcreate(file, "(4-byte) integers", H5T_STD_I32LE, dspace, H5P_DEFAULT, dcpl, H5P_DEFAULT)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_dset; } // grow from here! // "add one row" if (H5Dset_extent(dset, (hsize_t[]){1}) < 0) { retval = EXIT_FAILURE; goto fail_extent; } // "add 99 more rows" // 100 = 1 + 99 if (H5Dset_extent(dset, (hsize_t[]){100}) < 0) { retval = EXIT_FAILURE; goto fail_extent; } // you can also shrink the dataset... fail_extent: H5Dclose(dset); fail_dset: H5Pclose(dcpl); fail_dcpl: H5Sclose(dspace); fail_dspace: H5Fclose(file); fail_file: return retval; }
Clinic 2021-08-17
Your Questions
- Q
- Will the HDF5 1.12.1 file locking changes be brought to 1.10.8?
- Q
- Are there or should there be special considerations when preserving HDF-5
files for future use? I support a research data repository at University of
Michigan and we occasionally receive these files (also netCDF and HDF-5
created by MATLAB).
- HDF5 feature use
- Relative paths, hard-coded paths (e.g., in external links)
- Dependencies such as plugins
- Metadata
- Faceted search, catalog, digest
- Check sums
- TODO: Create some guidance!
- HDF5 feature use
Last week's highlights
- Announcements
- New compression plugin based on Snappy-CUDA
- A new (de-)compression filter plugin using Google's Snappy algorithm
- It runs on GPUs
- By Mr. HDF5-UDF (Lucas C. Villa Real)
- New compression plugin based on Snappy-CUDA
- Forum
- Alignment of Direct Write Chunks
- Store large 1D datasets across multiple HDF5 file
- Receive compressed chunks w/ fixed number of samples/chunk
- Want to use direct chunk write
- Problem: Chunks may contain boundary chunks containing samples that belong to different datasets in different files
- Sub-optimal solution: Decompress the chunk, separate the samples, & use some kind of masking value on the next dataset
- Better solution?
- Alignment of Direct Write Chunks
Tips, tricks, & insights
- Virtual Datasets (VDS)
- Logically, HDF5 datasets have a shape (rank or dimensionality) and an element type
- Physically, HDF5 datasets have a layout (in a logical HDF5 file): contiguous, chunked, compact, virtual
- A virtual dataset is an HDF5 dataset of virtual layout (- duh!)
- Virtual layout: some or all of the dataset's elements are stored in constituent datasets in the same or other HDF5 files, including other virtual datasets(!)
- Like any HDF5 dataset, HDF5 datasets of virtual layout have a shape (a.k.a. dataspace) and an element type (a.k.a datatype)
- Virtual datasets are constructed by specifying how selections(!) on constituent datasets map to regions in the virtual dataset's dataspace
Main API call:
H5Pset_virtual1: 2: herr_t H5Pset_virtual(hid_t vds_dcpl_id, // VDS creation properties 3: hid_t vds_dspace_id, // VDS dataspace 4: const char* src_file_name, // source file path 5: const char* src_dset_name, // source dataset path 6: hid_t src_space_id); // source dataspace select. 7:
- Sometimes multiple calls to
H5Pset_virtualare necessary, but there's support forprintf-style format strings to describe multiple source files & datasets - Typically, a VDS is just a piece of (HDF5-)metadata
- How does that lead to a better solution? Use VDS to correct for data acquisition artifacts!
- Two approaches
- Write the "boundary chunk" to both datasets/files
- Write the "boundary chunk" to only one dataset/file
- In either case, we use VDS as a mechanism to construct the correct (time-delineated) datasets
- Main practical differences between 1. and 2.:
- Unless the data is WORM (write-once/read-many), there is a potential coherence problem in 1. because we have two copies of the halo data
- When accessing a dataset whose boundary chunk ended up in another file, under 2., the HDF5 library has to open another file and dataset, and locate the chunk
- The canonical VDS reference is RFC: HDF5 Virtual Dataset
- Good source of use cases and examples
- Not everything described in the RFC was implemented, e.g., datatype conversion
h5pyhas a nice interface for VDS
Clinic 2021-08-10
Your Questions
- Q
- Will the HDF5 1.12.1 file locking changes be brought to 1.10.8?
Last week's highlights
- Announcements
- Forum
- Problem compiling HDF5 using CMake with
HDF5_ENABLE_PARALLEL- Affects HDF5 1.12.1
- Discovered by Jan-Willem Blokland & turns out to be a goof-up in our CMake files
- Fixed in PR #843
- Problem compiling HDF5 using CMake with
Tips, tricks, & insights
- What is SWMR & what's new w/ VFD SWMR?
- SWMR = Single Writer Multiple Readers
- Use case: "Process collaboration w/o communication"
- Read from an HDF5 file that is actively being written
- "w/o communication" = no inter-process communication (IPC) required
- That's a big ask!
- How do we ensure that the readers don't read invalid, inconsistent, or corrupt data?
- How do we ensure that readers eventually see updates?
- Can we bound that delay?
- Does this require any special HW/SW support?
- Initial release in HDF5 1.10.0 (March 30 2016)
- Limitations of the first implementation
- No support for new items, e.g., objects, attributes, etc., no deletion
- Dataset append only
- Reliance on strict write ordering and atomic write guarantee as per POSIX semantics
- Many file systems don't do that, e.g., NFS
- Implementation touches most parts of the HDF5 library: high maintenance cost
- No support for new items, e.g., objects, attributes, etc., no deletion
- What VFD SWMR brings
- Arbitrary item and object creation/deletion
- Configurable bound (maximum time) between write and read
- Easier to maintain because of VFD-level implementation
- Relaxed storage requirements, i.e., the implementation can be modified to support NFS or object stores
- How is it done?
- Writer generates periodic snapshots of metadata at points when it's known to be in a consistent state
- These snapshot live outside the HDF5 file proper
- Readers' MD requests are satisfied from snapshots or unchanged MD in the HDF5 file
- Devil's in the detail, e.g., to guarantee time between write and read, we need to bound the maximum size of MD changes and use page buffering
- See the RFC for the details
- Writer generates periodic snapshots of metadata at points when it's known to be in a consistent state
Clinic 2021-08-03
Your Questions
- Q
- Will the HDF5 1.12.1 file locking changes be brought to 1.10.8?
- Q
- I’m interested in PyDarshan and its analysis of HDF5 Darshan Logs. The current resource that I have is this. Any other reference or documentation that you could point out? Thank you (Marta Garcia, ANL)
Last week's highlights
- Announcements
- HDFView 3.1.3 was released
- HDFView 3.1.3 was released
- Forum
- Check all members of compound datatype are present?
- Error is in the eye of the beholder
H5Tequalis of limited use- Provide or modify symbolic information in a single place
- Use named datatypes for documentation
- pHDF5 1.12: data transform in combination with compression filter
- Poor documentation (on our part)
- Who reads release notes?
- Dropped feature in "rush to the release"
- Will add a note to
H5Pset_compressionXcalls
- Poor documentation (on our part)
- Check all members of compound datatype are present?
Tips, tricks, & insights
- New function
H5Dchunk_iter
- Lets you iterate over dataset chunks, for example, to explore variability in compression
- Currently in the
developbranch
Let's write a simple "chunk analyzer!"
- Basic idea
Provide an HDF5 file name as the single argument.
#include "hdf5.h" #include <stdlib.h> #include <stdio.h> static herr_t visit_cb(hid_t obj, const char *name, const H5O_info2_t *info, void *op_data); int main(int argc, char **argv) { int retval = EXIT_SUCCESS; hid_t file; char path[] = {"/"}; if (argc < 2) { printf("HDF5 file name required!"); return EXIT_FAILURE; } if ((file = H5Fopen(argv[1], H5F_ACC_RDONLY, H5P_DEFAULT)) == H5I_INVALID_HID) { retval = EXIT_FAILURE; goto fail_file; } // let's visit all objects in the file if (H5Ovisit(file, H5_INDEX_NAME , H5_ITER_NATIVE , &visit_cb, path, H5O_INFO_BASIC) < 0) { retval = EXIT_FAILURE; goto fail_visit; } fail_visit: H5Fclose(file); fail_file: return retval; }
- Callback for
H5Ovisit
static int chunk_cb(const hsize_t *offset, uint32_t filter_mask, haddr_t addr, uint32_t nbytes, void *op_data); herr_t visit_cb(hid_t obj, const char *name, const H5O_info2_t *info, void *op_data) { herr_t retval = 0; char* base_path = (char*) op_data; if (info->type == H5O_TYPE_DATASET) // current object is a dataset { hid_t dset, dcpl; if ((dset = H5Dopen(obj, name, H5P_DEFAULT)) == H5I_INVALID_HID) { retval = -1; goto func_leave; } if ((dcpl = H5Dget_create_plist(dset)) == H5I_INVALID_HID) { retval = -1; goto fail_dcpl; } if (H5Pget_layout(dcpl) == H5D_CHUNKED) // dataset is chunked { __label__ fail_dtype, fail_dspace, fail_fig; hid_t dspace, dtype; size_t size, i; int rank; hsize_t cdims[H5S_MAX_RANK]; // get resources if ((dtype = H5Dget_type(dset)) < 0) { retval = -1; goto fail_dtype; } if ((dspace = H5Dget_space(dset)) < 0) { retval = -1; goto fail_dspace; } // get the figures if ((size = H5Tget_size(dtype)) == 0 || (rank = H5Sget_simple_extent_ndims(dspace)) < 0 || H5Pget_chunk(dcpl, H5S_MAX_RANK, cdims) < 0) { retval = -1; goto fail_fig; } // calculate the nominal chunk size size = 1; for (i = 0; i < (size_t) rank; ++i) size *= cdims[i]; // print dataset info printf("%s%s : nominal chunk size %lu [B] \n", base_path, name, size); // get the allocated chunk sizes if (H5Dchunk_iter(dset, H5P_DEFAULT, &chunk_cb, NULL) < 0) { retval = -1; goto fail_fig; } fail_fig: H5Sclose(dspace); fail_dspace: H5Tclose(dtype); fail_dtype:; } H5Pclose(dcpl); fail_dcpl: H5Dclose(dset); } func_leave: return retval; }
- Callback for
H5Dchunk_iter
int chunk_cb(const hsize_t *offset, uint32_t filter_mask, haddr_t addr, uint32_t nbytes, void *op_data) { // for now we care only about the allocated chunk size printf("%d\n", nbytes); return EXIT_SUCCESS; }
Clinic 2021-07-27
Your Questions
- Q
- Will the HDF5 1.12.1 file locking changes be brought to 1.10.8?
- Q
- Is there any experience using HDF5 for MPI output with compression on a progressively striped Lustre system? We’re seeing some file corruption and we are wondering where the problem lies. - Sean Freeman
- A
- Nothing comes to mind that's related to that, but it might be good to
see what MPI and MPI I/O backend the user is using, since we've had issues
with ROMIO in the past for example. - Jordan Henderson
- HPE MPT from SGI, not using ROMIO
- Maybe an MVE?
Last week's highlights
- Announcements
- 2021 HDF5 User Group Call for Papers
- Format: "Paper and Presentation" or "Presentation"
- Deadline for presentation abstracts:
- Event: 12-15 October 2021
- HDF5 RFCs
- All RFCs in one place
- 2021 HDF5 User Group Call for Papers
- Forum
- H5idecref hangs
- Parallel HDF5 "misbehavior" can be subtle
- Collective Calling Requirements in Parallel HDF5 Applications
- Allocation time, fill behavior, etc., are less visible, but included
- Always ask if an operation has the potential to create any "metadata disparity?"
- How to read a UTF-8 string
H5T_CSET_UTF8for character encoding- The is currently a subtle bug in string handling Inconsistent behaviour with variable-length strings and character encoding #544
- ASCII is a proper subset of UTF-8 with byte-identical encodings
- The HDF5 library is blissfully ignorant about that…
- When should MPI barrier be used when using Parallel HDF5?
- Only if you want to achieve a certain consistency semantics ("processes communicating through a file")
- Make a wish!
- What small changes would make a big difference in your HDF5 workflow?
- Chime in!
- H5idecref hangs
Tips, tricks, & insights
- User-defined Properties
- Use case: You want to pass property list-like things (dictionaries) around,
your language doesn't have dictionaries, and you don't want to re-invent the
wheel
- You want to stay close to the "HDF5 way of doing things"
- See General Property List Operations (Advanced)
- You can define your own property list classes w/ pre-defined or "permanent" properties
- You can insert "temporary" (= non-permanent) properties into any property list
- WARNING: Permanent or temporary, none of this is persisted in the HDF5
file!
- These property lists (and properties) get copied between APIs provided you've implemented the necessary callbacks
- Depending on the property value types, make sure you implement proper resource management, or memory leaks might occur
- It's an esoteric/advanced/infrequently used feature, but might be just what you need in certain circumstances
- Use case: You want to pass property list-like things (dictionaries) around,
your language doesn't have dictionaries, and you don't want to re-invent the
wheel
Clinic 2021-07-20
Your Questions
- Q
- Will the HDF5 1.12.1 file locking changes be brought to 1.10.8?
- Q
- Named types, what are the benefits?
- A
- Documentation and convenience. You don't have to (re-)create the datatype over and over. Just open it and pass the handle to attribute and dataset creations!
Last week's highlights
- Announcements
- 2021 HDF5 User Group Call for Papers
- Format: "Paper and Presentation" or "Presentation"
- Deadline for presentation abstracts:
- Event: 12-15 October 2021
- 2021 HDF5 User Group Call for Papers
- Forum
- Migrating pandas and local HF5 to HSDS
- John Readey posted a nice comment referencing an article that shows how to map
pandas dataframes to HDF5 via
h5py - The same will also work w/
h5pyd
- John Readey posted a nice comment referencing an article that shows how to map
pandas dataframes to HDF5 via
- Local HSDS performance vs local HDF5 files
- Interesting exchange of benchmark results
- Data (response) preparation in HSDS seems to be slow
- The big question is why HSDS is sending data at a 10x lower rate than a vanilla REST API (339 MB/s versus 4,384 MB/s)
- MPI-IO file info actually used
- The
MPI_Infoobject returned byH5Pget_fapl_mpiodoes not return the full set of hints seen by MPI
- The
- Make a wish!
- What small changes would make a big difference in your HDF5 workflow?
- Chime in!
- Migrating pandas and local HF5 to HSDS
Tips, tricks, & insights
- HDF5 File Images
- Use cases
- In-memory I/O
- Share HDF5 data between processes w/o a file system
- Transmit HDF5 data packets over a network
- See also Vijay Kartik's (DESY) presentation and slides from HUG 2021 Europe
- Starting point: HDF5 core VFD
- Replace the file (logical byte sequence) with a memory buffer
read,write->memcpy
- HDF5 file images generalize that concept
- Memory buffer + management interface (callbacks)
- Reference: HDF5 File Image Operations
- HDF5 file images can be exchanged between processes via IPC (shared memory segment) or a TCP connection
See section 4 (Examples) in the reference
+++ Process A +++ +++ Process B +++ <Open and construct the desired file hid_t file_; with the Core file driver> H5Fflush(fid); size = H5Fget_file_image(fid, NULL, 0); buffer_ptr = malloc(size); H5Fget_file_image(fid, buffer_ptr, size); <transmit size> <receive size> buffer_ptr = malloc(size) <transmit *buffer_ptr> <receive image in *buffer_ptr> free(buffer_ptr); <close core file> file_id = H5LTopen_file_image ( buf, buf_len, H5LT_FILE_IMAGE_DONT_COPY ); <read data from file, then close. note that the Core file driver will discard the buffer on close>
- Use cases
Clinic 2021-07-13
Your Questions
- Q
- Will the HDF5 1.12.1 file locking changes be brought to 1.10.8?
Last week's highlights
- HUG 2021 Europe
- The presentations from the European HUG 2021 event are now available on YouTube
- Announcements
- 2021 HDF5 User Group Call for Papers
- Format: "Paper and Presentation" or "Presentation"
- Deadline for presentation abstracts:
- Event: 12-15 October 2021
- 2021 HDF5 User Group Call for Papers
- Forum
- HDF5 specs unclear: B-tree v2 and fractal heap
- There was a lot of interest in accessing HDF5 files directly, i.e., w/o the HDF5 library
- Good clarity test for the file format spec.
- Someone is extending the
pyfivepure Python HDF5 reader and asked for heap ID clarification
- Building REST VOL on Windows 10
- REST VOL = access HSDS via the HDF5 library, converts HDF5 library calls into RESTful requests (and back)
- Requires
libcurland massaging the linker flags
- File storage on AWS S3
- Where did the data go?
- The documentation can be found here
- Migrating pandas and local HF5 to HSDS
- Using DigitalOcean and spaces instead of AWS S3
- Using
pandas- Will that work?
- Local HSDS performance vs local HDF5 files
- User observed a 10x slowdown for reads
- What to expect?
- Several sources for slowdown (e.g., compression, IPC)
- John Ready is working on a comprehensive performance test suite
- Start list of VOL plugins?
- h5diff and ignoring attributes
- Now in HDF5
developas--exclude-attributeoption forh5diff
- Now in HDF5
- Make a wish!
- What small changes would make a big difference in your HDF5 workflow?
- Chime in!
- HDF5 specs unclear: B-tree v2 and fractal heap
Clinic 2021-07-06
Your Questions
- Q
- Will the HDF5 1.12.1 file locking changes be brought to 1.10.8?
Last week's highlights
- Announcements
- European HDF Users Group (HUG) Summer 2021 July 7-8, 2021
- 2021 HDF5 User Group Call for Papers
- Format: "Paper and Presentation" or "Presentation"
- Deadline for presentation abstracts:
- Event: 12-15 October 2021
- Reference Manual in Doxygen
- New permanent home for HDF5 documentation
- https://docs.hdfgroup.org/hdf5/develop/
- https://docs.hdfgroup.org/hdf5/v112/
- European HDF Users Group (HUG) Summer 2021 July 7-8, 2021
- Forum
- Make a wish!
- What small changes would make a big difference in your HDF5 workflow?
- Chime in!
- Problems with HDF5 and SWMR on NFS mounted network disk
- Current SWMR implementation depends on POSIX write semantics (esp. order preservation)
- NFS & co. don't do that
- VFD SWMR might change that
- Save related data
- Make a wish!
Tips, tricks, & insights
- HDF5 ecosystem: HDFql
- HDFql = Hierarchical Data Format query language
- High-level and declarative
- SQL is the gold standard for simplicity and power
- Adapted to HDF5
- A single guest language (HDFql) for multiple host languages (C, C++, Java, Python, C#, Fortran, R)
- Seamless parallelism (multiple cores, MPI)
- Example
- Host language: Fortran
- Find all datasets existing in an HDF5 file named
data.h5that start withtemperatureand are of data typefloat - For each dataset found, print its name and read its data
- Write the data into a file named
output.txtin an ascending order - Each value (belonging to the data) is written in a new line using a UNIX-based end of line (EOL) terminator
PROGRAM Example USE HDFql INTEGER :: state state = hdfql_execute("USE FILE data.h5") state = hdfql_execute( "SHOW DATASET LIKE **/^temperature WHERE DATA TYPE == FLOAT") D O WHILE(hdfql_cursor_next() .EQ. HDFQL_SUCCESS) WRITE(*, *) "Dataset found: ", hdfql_cursor_get_char() state = hdfql_execute( "SELECT FROM " // hdfql_cursor_get_char() // " ORDER ASC INTO UNIX FILE output.txt SPLIT 1") END DO state = hdfql_execute("CLOSE FILE") END PROGRAMCREATE FILE my_file.h5 CREATE FILE experiment.h5 IN PARALLEL CREATE GROUP countries CREATE DATASET values AS FLOAT(20, 40) ENABLE ZLIB INSERT INTO measurements VALUES FROM EXCEL FILE values.xlsx INSERT INTO dset(0:::1) VALUES FROM MEMORY 0 SHOW ATTRIBUTE group2 LIKE **/1|3
- Links
- HDFql home
- Binaries for Linux, Windows, and macOS
- HDFql examples
- HDFql license
- HDFql home
Clinic 2021-06-29
Your Questions
- Q
- Will the HDF5 1.12.1 file locking changes be brought to 1.10.8?
Last week's highlights
- Announcements
- European HDF Users Group (HUG) Summer 2021 July 7-8, 2021
- Propose a presentation–let us know you’re interested as soon as possible.
- Finalize your presentation by sending in your abstract by June 30
- Submit your PPT and other materials by July 6
- Attendee and Speaker Registration
- 2021 HDF5 User Group Call for Papers
- Format: "Paper and Presentation" or "Presentation"
- Deadline for presentation abstracts:
- Event: 12-15 October 2021
- European HDF Users Group (HUG) Summer 2021 July 7-8, 2021
- Forum
- Make a wish!
- What small changes would make a big difference in your HDF5 workflow?
- Chime in!
- HDF5 Performance
- User is seeing decent performance w/ packet tables
H5PT - Issue w/ deleting links (?)
- User is seeing decent performance w/ packet tables
- Local HSDS performance vs local HDF5 files
- HDFView should default to read-only open
- Yes!
- Too many options? Sloppy testing?
- HDF - port to Rust
- Not a bad idea: a BIG job
- Perhaps not a replacement for the C implementation (How many systems have a Rust compiler?)
- I understand that that's not the point, but there are reasonably mature Rust language bindings for HDF5
- Make a wish!
Tips, tricks, & insights
- How do I delete an HDF5 item?
- HDF5 item = something a user created and that gets stored in an HDF5 file
- High-level view
- Attributes -
H5Adelete* - Objects (groups, datasets, named datatypes) -
H5Ldelete*
- Attributes -
- Low-level view
- Objects are reference-counted (in the object OHDR in the file!)
- A positive reference count means the object is considered in-use or referenced
- A zero reference count signals to the HDF5 library free space availability
- If that free space can be used or reclaimed depends on several factors
- Position of the gap (middle of the file, end of the file)
- Intervening file closure
- Library version free-space management and tracking support
- Virtual File Driver support
- A detailed description of file space management (including free space) can be found in this RFC
- Highlights:
- Pre-HDF5 1.10.x
- Free space info is not persisted across file open/close epochs
- Typical symptom: deleting an object in another epoch will not reduce file size
- Use
h5statto discover the amount of free-/unused space h5repackis the cheapest way to recover unused space- May not be practical for large files
- Free space info is not persisted across file open/close epochs
- HDF5 1.10.x+
- Free space info can be persisted across file open/close epochs
- Needs to be enabled in file creation property list
- Set threshold on smallest quanta to be tracked
- Combine with paged allocation!
- Free space info can be persisted across file open/close epochs
- Pre-HDF5 1.10.x
- The story too involved for most users
- Summary
- Don't create (in the file) what you don't need
- Use
h5statto assess andh5repackto reclaim free space: don't obsess over a few KB! - If you really want to get into file space management, use HDF5 1.10.x+ and come back next time with a question!
Clinic 2021-06-22
Your Questions
- Q
- What is the CacheVOL and what can I do with it? How can I use node-local
storage on an HPC system?
- Complexity is hidden from users
- Use in conjunction w/ Async VOL
- Data migration to and from the remote storage is performed in the background
- Developed by NERSC w/ Huihuo Zheng as the lead developer
- No official release yet
- See this ECP BoF presentation (around slide 29)
- GitHub
- Spack integration
- Q
- Will the HDF5 1.12.1 file locking changes be brought to 1.10.8?
Last week's highlights
- Announcements
- European HDF Users Group (HUG) Summer 2021 July 7-8, 2021
- Propose a presentation–let us know you’re interested as soon as possible.
- Finalize your presentation by sending in your abstract by June 30
- Submit your PPT and other materials by July 6
- Attendee and Speaker Registration
- 2021 HDF5 User Group Call for Papers
- Format: "Paper and Presentation" or "Presentation"
- Deadline for presentation abstracts:
- Event: 12-15 October 2021
- HDF5-UDF 2.1 released – with support for Windows and macOS!
- European HDF Users Group (HUG) Summer 2021 July 7-8, 2021
- Forum
- Make a wish!
- What small changes would make a big difference in your HDF5 workflow?
- Chime in!
- HDF5 Performance
- Slowdown w/ a lot of attributes (10K, 100K, …)
- Slight improvement w/ a low-level fix (see below)
- The customer is always right, but maybe this is not such a good idea
- Make a wish!
Tips, tricks, & insights
- How do I use a newer HDF5 file format?
- Versions
- HDF5 library
- File format specification
- HDF5 library forward- and backward-compatibility
- Backward
- The latest version of the library can read HDF5 files created with all earlier library versions
- Forward
- A given version of the library can read all (objects in) HDF5 files created by later versions as long as they are compatible with this version.
- By default, newer HDF5 library versions use settings compatible with the earliest library version
#include "hdf5.h" #include <stdio.h> #include <stdlib.h> int main() { __label__ fail_fapl, fail_file; int ret_val = EXIT_SUCCESS; hid_t fapl, file; { unsigned maj, min, rel; if (H5get_libversion(&maj, &min, &rel) < 0) { ret_val = EXIT_FAILURE; goto fail_fapl; } printf("Welcome to HDF5 %d.%d.%d!\n", maj, min, rel); } if ((fapl = H5Pcreate(H5P_FILE_ACCESS)) < 0) { ret_val = EXIT_FAILURE; goto fail_fapl; } // bracket the range of LIBRARY VERSIONS for object creation and access, // e.g., min. vers. 1.8, max. version current if (H5Pset_libver_bounds(fapl, H5F_LIBVER_V18, H5F_LIBVER_LATEST) < 0) { ret_val = EXIT_FAILURE; goto fail_file; } if ((file = H5Fcreate("my.h5", H5F_ACC_TRUNC, H5P_DEFAULT, fapl)) < 0) { ret_val = EXIT_FAILURE; goto fail_file; } // do something useful w/ FILE H5Fclose(file); fail_file: H5Pclose(fapl); fail_fapl:; return ret_val; }
- Versions
Clinic 2021-06-15
Your Questions
- CacheVOL
- How to use local storage?
- Hermes webinar
Last week's highlights
- Announcements
- 2021 HDF5 User Group Call for Papers
- Format: "Paper and Presentation" or "Presentation"
- Deadline for presentation abstracts:
- Event: 12-15 October 2021
- HDF5 1.12.1-6-rc2 source available for testing
- You can help!
- 2021 HDF5 User Group Call for Papers
- Forum
- Make a wish!
- What small changes would make a big difference in your HDF5 workflow?
- Chime in!
- Issue unlocking HDF5 file?
- Access to ‘miscellaneous dataset information’/get size of compressed dataset
Aleksandar's
h5pyexample:import h5py def comp_ratio(name, obj): if isinstance(obj, h5py.Dataset) and obj.chunks is not None: dcpl = obj.id.get_create_plist() if dcpl.get_nfilters(): stor_size = obj.id.get_storage_size() if stor_size != 0: ratio = float(obj.nbytes) / float(stor_size) print(f'Compression ratio for "{obj.name}": {ratio}') fname = 'example.h5' with h5py.File(fname, mode='r') as h5f: h5f.visititems(comp_ratio)
- Make a wish!
Tips, tricks, & insights
- File Locking (Dana Robinson)
- Outline
The basic file locking algorithm is simple:
- On opening the file, we place the lock as described below. This is true for all file opens, not just SWMR (Single Write Multiple Readers).
- For SWMR writers, this lock is removed after we flush the file's superblock.
- All other processes will hold the lock until the file is closed or
H5Fstart_swmr_write()is called.
- Architecture
File locking is handled in the native HDF5 virtual object layer (VOL) connector, so other VOL connectors (REST, etc.) don't do any locking.
File locking is handled at the library level, not the virtual file level (VFL). Virtual file drivers (VFDs) do have to provide an implementation of the lock and unlock VFD operations for file locking to work, though. If a VFD doesn't provide a lock operation, file locking will be ignored when using that VFD. Most of the VFDs provided with the library are based on the POSIX SEC2 VFD (the default on all platforms, including Windows) and provide the locking I've described.
The stdio VFD only uses
flock(2)when it's available, it ignores file locking when it's not (e.g., on Windows). This is because the stdio VFD is a demo VFD that uses very little of the library's helper functions and macros and that's where the flock/fcntl/fail code lies.The MPI-IO VFD, as you might expect, ignores file locking.
- SWMR
The
H5Fstart_swmr_write()API call will unlock the file after it flushes everything in memory.Related to the OS-level locking algorithm, if the file was opened by a SWMR writer (either by using the
H5F_ACC_SWMR_WRITEflag at create/open or viaH5Fstart_swmr_write()) it will have its superblock marked as such. This mark will prevent readers from opening the file unless they open it with theH5F_ACC_SWMR_READflag.HDF5 1.8.x and earlier do not understand this version of the superblock and will return an error code when trying to open the file. This mark is cleared when the file is closed. If the writer crashes, you can remove the mark using the
h5cleartool provided with the library. - UNIX/Linux, Non-Windows
Compile time option:
--enable-file-locking=(yes|no|best-effort) Sets the default for whether or not to use file locking when opening files. Can be overridden with the HDF5_USE_FILE_LOCKING environment variable and the H5Pset_file_locking() API call. best-effort attempts to use file locking but does not fail when file locks have been disabled on the file system (useful with Lustre). [default=best-effort]You can disable all file locking at runtime by setting an environment variable named
HDF5_USE_FILE_LOCKINGto the string"FALSE".We preferentially use
flock(2)in POSIX-like environments where it's available. If that is not available, we fall back onfcntl(2). If that is not found and not best effort, the lock operation uses an internal function that simply fails.With
flock(2), we useLOCK_EXwith read/write permissions andLOCK_SHwith read-only. Both are combined withLOCK_NBto create non-blocking locks.With
fcntl(2), we lock the entire file. We useF_WRLCKwith read/write permissions andF_RDLCKwith read-only. - Windows
There is no locking on Windows systems since the Windows POSIX layer doesn't support that. File locking on Windows is just a no-op (as opposed to failing, as we do when neither
flock(2)norfcntl(2)are found). We'd need a a virtual file driver based on Win32 API calls to handle file locking on Windows.Windows uses the POSIX VFD as the default driver. We do not (yet) have a VFD that uses Win32 API calls like
CreateFile(). The POSIX layer in Windows is incomplete, however, and does not includeflock(2)orfcntl(2)so we simply skip file locking there for the time being.See below for an update!
- Summary
File locking is only implemented to help prevent users from accessing files when SWMR write ordering is not turned on (or when we're doing the superblock marking). It's not inherent to the SWMR algorithm, which is lock-free and instead based on write ordering.
- Hot off the press
In the 1.12.1-6-rc2 release notes, we find this entry:
• File locking updates: File locks now work on Windows Adds BEST_EFFORT value to HDF5_USE_FILE_LOCKING environment variable Adds H5Pset/get_file_locking() API calls Adds --enable-file-locking=(yes|no|best-effort) option to Autotools Adds HDF5_USE_FILE_LOCKING and HDF5_IGNORE_DISABLED_FILE_LOCKS to CMake
- Outline
Clinic 2021-06-08
Your Questions
???
Last week's highlights
- Announcements
- 2021 HDF5 User Group Call for Papers
- Format: "Paper and Presentation" or "Presentation"
- Deadline for presentation abstracts:
- Event: 12-15 October 2021
- 2021 HDF5 User Group Call for Papers
- Forum
- Make a wish!
- What small changes would make a big difference in your HDF5 workflow?
- Great comments already
- Revised filter interface
- Updates to
HDF5_PLUGIN_PATH - Amalgamated source
- Modern language bindings for Fortran
- Chime in!
- Issue unlocking HDF5 file?
- Case of poor documentation & flip-flopping on our part?
H5I_dec_refhangs
- Make a wish!
Tips, tricks, & insights
- Jam-packed HDF5 Files - The HDF5 User Block
- "Keeping things together." - mantra
- Metadata and data
- Stuff - a zip file of ancillary (non-HDF5) data, documentation, etc.
- "HDF5 can be on the inside or the outside"
- Reserved space at the beginning of an HDF5 file
- Fixed size 2N bytes, min. size 512 KiB
- Ignored by the HDF5 library
Tooling
h5jam,h5unjamusage: h5jam -i <in_file.h5> -u <in_user_file> [-o <out_file.h5>] [--clobber] Adds user block to front of an HDF5 file and creates a new concatenated file. OPTIONS -i in_file.h5 Specifies the input HDF5 file. -u in_user_file Specifies the file to be inserted into the user block. Can be any file format except an HDF5 format. -o out_file.h5 Specifies the output HDF5 file. If not specified, the user block will be concatenated in place to the input HDF5 file. --clobber Wipes out any existing user block before concatenating the given user block. The size of the new user block will be the larger of; - the size of existing user block in the input HDF5 file - the size of user block required by new input user file (size = 512 x 2N, N is positive integer.) -h Prints a usage message and exits. -V Prints the HDF5 library version and exits. Exit Status: 0 Succeeded. >0 An error occurred.usage: h5unjam -i <in_file.h5> [-o <out_file.h5> ] [-u <out_user_file> | --delete] Splits user file and HDF5 file into two files: user block data and HDF5 data. OPTIONS -i in_file.h5 Specifies the HDF5 as input. If the input HDF5 file contains no user block, exit with an error message. -o out_file.h5 Specifies output HDF5 file without a user block. If not specified, the user block will be removed from the input HDF5 file. -u out_user_file Specifies the output file containing the data from the user block. Cannot be used with --delete option. --delete Remove the user block from the input HDF5 file. The content of the user block is discarded. Cannot be used with the -u option. -h Prints a usage message and exits. -V Prints the HDF5 library version and exits. If neither --delete nor -u is specified, the user block from the input file will be displayed to stdout. Exit Status: 0 Succeeded. >0 An error occurred.- Let's try this!
- "Keeping things together." - mantra
Clinic 2021-06-01
Your Questions
- Does
h5repackhave any impact on reading?- What can
h5repackdo for you?- Reclaim unused file space
- (Down-)Upgrade file format features
- Change dataset layout
- (Un-)Compress datasets
- … (incomplete list! - Run
h5dump --help!)
- Yes, the read performance of a re-packed HDF5 file could be better or worse (or about the same).
- What can
- Is there any difference in reading a variable/field if it is compressed or
un-compressed? (This question came in at the end of our May 18 session.)
- Assuming loss-less compression, no, in terms of value
- Yes, most likely, because (de-)compression requires CPU-cycles
- Potential reduction in I/O bandwidth
- Pathology: the data size increases as a result of compression
- HDF5 Data Flow Pipeline for
H5Dread
- Do you have recommendations for setting Figure of Merit (FOM) to
measure/capture I/O improvements? Any consideration based on current
supercomputers/hybrid systems, # of files used, kind of I/O (e.g. different
for read than for write), HDF5 versions, HDF5 features, if using SSDs/Burst
buffers, etc. What would be a good sample of FOM to follow?
- Baseline, metric (file size, throughput, IOPs)
- Large number of combinations? Perhaps polar diagrams? See this webinar around 15:18.
Last week's highlights
- Announcements
- 2021 HDF5 User Group Call for Papers
- Format: "Paper and Presentation" or "Presentation"
- Deadline for abstracts: 1 June 2021
- Event: 12-15 October 2021
- 2021 HDF5 User Group Call for Papers
- Forum
- HDF5-UDF runs on macOS!
- Caveat: Apple's Operating Systems Are Malware says FSF
- Make a wish!
- What small changes would make a big difference in your HDF5 workflow?
- HDF5-UDF runs on macOS!
Tips, tricks, & insights
- Jam-packed HDF5 Files - The HDF5 User Block
- "Keeping things together." - mantra
- Metadata and data
- Stuff - a zip file of ancillary (non-HDF5) data, documentation, etc.
- "HDF5 can be on the inside or the outside"
- Reserved space at the beginning of an HDF5 file
- Fixed size 2N bytes, min. size 512 KiB
- Ignored by the HDF5 library
Tooling
h5jam,h5unjamusage: h5jam -i <in_file.h5> -u <in_user_file> [-o <out_file.h5>] [--clobber] Adds user block to front of an HDF5 file and creates a new concatenated file. OPTIONS -i in_file.h5 Specifies the input HDF5 file. -u in_user_file Specifies the file to be inserted into the user block. Can be any file format except an HDF5 format. -o out_file.h5 Specifies the output HDF5 file. If not specified, the user block will be concatenated in place to the input HDF5 file. --clobber Wipes out any existing user block before concatenating the given user block. The size of the new user block will be the larger of; - the size of existing user block in the input HDF5 file - the size of user block required by new input user file (size = 512 x 2N, N is positive integer.) -h Prints a usage message and exits. -V Prints the HDF5 library version and exits. Exit Status: 0 Succeeded. >0 An error occurred.usage: h5unjam -i <in_file.h5> [-o <out_file.h5> ] [-u <out_user_file> | --delete] Splits user file and HDF5 file into two files: user block data and HDF5 data. OPTIONS -i in_file.h5 Specifies the HDF5 as input. If the input HDF5 file contains no user block, exit with an error message. -o out_file.h5 Specifies output HDF5 file without a user block. If not specified, the user block will be removed from the input HDF5 file. -u out_user_file Specifies the output file containing the data from the user block. Cannot be used with --delete option. --delete Remove the user block from the input HDF5 file. The content of the user block is discarded. Cannot be used with the -u option. -h Prints a usage message and exits. -V Prints the HDF5 library version and exits. If neither --delete nor -u is specified, the user block from the input file will be displayed to stdout. Exit Status: 0 Succeeded. >0 An error occurred.- Let's try this!
- "Keeping things together." - mantra
Clinic 2021-05-25
Your Questions
- Does
h5repackhave any impact on reading?- Yes, the read performance of a re-packed HDF5 file could be better or worse (or about the same).
- Is there any difference in reading a variable/field if it is compressed or un-compressed? (This question came in at the end of our May 18 session.)
- Do you have recommendations for setting Figure of Merit (FOM) to
measure/capture I/O improvements? Any consideration based on current
supercomputers/hybrid systems, # of files used, kind of I/O (e.g. different
for read than for write), HDF5 versions, HDF5 features, if using SSDs/Burst
buffers, etc. What would be a good sample of FOM to follow?
- Baseline, metric
- Large number of combinations? Perhaps polar diagrams? See this webinar around 15:18.
Last week's highlights
- Announcements
- The Hermes Buffering System - Take Two was POSTPONED
- Stay tuned for a re-announcement
- 2021 HDF5 User Group Call for Papers
- Format: "Paper and Presentation" or "Presentation"
- Deadline for abstracts: 1 June 2021
- Event: 12-15 October 2021
- The Hermes Buffering System - Take Two was POSTPONED
- Forum
Tips, tricks, & insights
h5repack- Getting stuff done w/o writing a lot of code
Sanity check:
h5repack --help
The output should look like this:
usage: h5repack [OPTIONS] file1 file2 file1 Input HDF5 File file2 Output HDF5 File OPTIONS -h, --help Print a usage message and exit -v, --verbose Verbose mode, print object information -V, --version Print version number and exit -n, --native Use a native HDF5 type when repacking --enable-error-stack Prints messages from the HDF5 error stack as they occur -L, --latest Use latest version of file format This option will take precedence over the options --low and --high --low=BOUND The low bound for library release versions to use when creating objects in the file (default is H5F_LIBVER_EARLIEST) --high=BOUND The high bound for library release versions to use when creating objects in the file (default is H5F_LIBVER_LATEST) --merge Follow external soft link recursively and merge data --prune Do not follow external soft links and remove link --merge --prune Follow external link, merge data and remove dangling link -c L1, --compact=L1 Maximum number of links in header messages -d L2, --indexed=L2 Minimum number of links in the indexed format -s S[:F], --ssize=S[:F] Shared object header message minimum size -m M, --minimum=M Do not apply the filter to datasets smaller than M -e E, --file=E Name of file E with the -f and -l options -u U, --ublock=U Name of file U with user block data to be added -b B, --block=B Size of user block to be added -M A, --metadata_block_size=A Metadata block size for H5Pset_meta_block_size -t T, --threshold=T Threshold value for H5Pset_alignment -a A, --alignment=A Alignment value for H5Pset_alignment -q Q, --sort_by=Q Sort groups and attributes by index Q -z Z, --sort_order=Z Sort groups and attributes by order Z -f FILT, --filter=FILT Filter type -l LAYT, --layout=LAYT Layout type -S FS_STRATEGY, --fs_strategy=FS_STRATEGY File space management strategy for H5Pset_file_space_strategy -P FS_PERSIST, --fs_persist=FS_PERSIST Persisting or not persisting free- space for H5Pset_file_space_strategy -T FS_THRESHOLD, --fs_threshold=FS_THRESHOLD Free-space section threshold for H5Pset_file_space_strategy -G FS_PAGESIZE, --fs_pagesize=FS_PAGESIZE File space page size for H5Pset_file_space_page_size ...There's a lot of stuff to chew over, but let's focus on the examples:
... Examples of use: 1) h5repack -v -f GZIP=1 file1 file2 GZIP compression with level 1 to all objects 2) h5repack -v -f dset1:SZIP=8,NN file1 file2 SZIP compression with 8 pixels per block and NN coding method to object dset1 3) h5repack -v -l dset1,dset2:CHUNK=20x10 -f dset3,dset4,dset5:NONE file1 file2 Chunked layout, with a layout size of 20x10, to objects dset1 and dset2 and remove filters to objects dset3, dset4, dset5 4) h5repack -L -c 10 -s 20:dtype file1 file2 Using latest file format with maximum compact group size of 10 and minimum shared datatype size of 20 5) h5repack -f SHUF -f GZIP=1 file1 file2 Add both filters SHUF and GZIP in this order to all datasets 6) h5repack -f UD=307,0,1,9 file1 file2 Add bzip2 filter to all datasets 7) h5repack --low=0 --high=1 file1 file2 Set low=H5F_LIBVER_EARLIEST and high=H5F_LIBVER_V18 via H5Pset_libver_bounds() when creating the repacked file, file2
Let's create some test data and play!
#include "hdf5.h" #include <assert.h> #include <stdlib.h> #define SIZE 1024*1024 int main() { int ret_val = EXIT_SUCCESS; hid_t file, fspace, dset; double* data = (double*) malloc(SIZE*sizeof(double)); if ((file = H5Fcreate("foo.h5", H5F_ACC_TRUNC, H5P_DEFAULT, H5P_DEFAULT)) == H5I_INVALID_HID) { ret_val = EXIT_FAILURE; goto fail_file; } if ((fspace = H5Screate_simple(1, (hsize_t[]){ SIZE }, NULL)) == H5I_INVALID_HID) { ret_val = EXIT_FAILURE; goto fail_fspace; } if ((dset = H5Dcreate(file, "sequential", H5T_IEEE_F64LE, fspace, H5P_DEFAULT, H5P_DEFAULT, H5P_DEFAULT)) == H5I_INVALID_HID) { ret_val = EXIT_FAILURE; goto fail_dset; } for (size_t i = 0; i < SIZE; ++i) data[i] = (double)i; if (H5Dwrite(dset, H5T_NATIVE_DOUBLE, H5S_ALL, H5S_ALL, H5P_DEFAULT, data) < 0) ret_val = EXIT_FAILURE; H5Dclose(dset); if ((dset = H5Dcreate(file, "random", H5T_IEEE_F64LE, fspace, H5P_DEFAULT, H5P_DEFAULT, H5P_DEFAULT)) == H5I_INVALID_HID) { ret_val = EXIT_FAILURE; goto fail_dset; } for (size_t i = 0; i < SIZE; ++i) data[i] = (double)rand()/(double)RAND_MAX; if (H5Dwrite(dset, H5T_NATIVE_DOUBLE, H5S_ALL, H5S_ALL, H5P_DEFAULT, data) < 0) ret_val = EXIT_FAILURE; H5Dclose(dset); fail_dset: H5Sclose(fspace); fail_fspace: H5Fclose(file); fail_file: free(data); assert(ret_val == EXIT_SUCCESS); return ret_val; }
Clinic 2021-05-18
Your Questions
???
Last week's highlights
- Announcements
- Recording of H5Coro: The HDF5 Cloud-Optimized Read-Only Library is available now
- The Hermes Buffering System - Take Two
- May 28, 2021 11:00 a.m. CDT
- Registration
- Previous Webinar Recording
- Hermes on GitHub
- 2021 HDF5 User Group Call for Papers
- Format: "Paper and Presentation" or "Presentation"
- Deadline for abstracts: 1 June 2021
- Event: 12-15 October 2021
- Forum
- HDF5-UDF 2.0 released: UDF signing, trust profiles, Python bindings, and more!
- UDF signing
- Library API
- Python bindings (i.e., manageable from Jupyter notebook)
- Source code storage
- GitHub
- Zlib-ng works with HDF5
- More anecdotal evidence of performance improvements
- Begs a maintenance question: Should we/ can we support multiple implementations of the same filter?
- Reference Manual in Doxygen
- Current version
- Life cycle entries - are they helpful?
- To be included in HDF5 1.12.1
- HDF5-UDF 2.0 released: UDF signing, trust profiles, Python bindings, and more!
Tips, tricks, & insights
- When should you consider using chunked layout for a dataset?
"Consider" means that you should also consider alternatives. None of the items listed below mandates chunked layout.
- Considerations
- I would like to use a compression or other filter w/ my data
- I cannot know/estimate the data size in advance
- I need the ability to append data indefinitely
- My read/write pattern is such that contiguous layout would reduce performance
- Caveats
- What's a good chunk size?
- Is my chunk cache the right size?
- Compound types?
- Variable-length datatypes?
- Are there edge chunks?
- Experimentation
- Don't waste your time writing a lot of code!
- Use a tool such as
h5repack - Use intuitive and boilerplate-free language bindings for Python, Julia, or C++ that exist thanks to the HDF community
- Use a tool such as
- Don't waste your time writing a lot of code!
- Considerations
Clinic 2021-05-11
Your Questions
- Where is the page that I'm showing?
- How did we prepare the webinar radial diagrams?
- Using Python's
plotlymodule - Here's a nice article to get you started.
- We'll include a few scripts w/ HDF5 I/O test in the future
- Using Python's
Last week's highlights
- Announcements
- H5Coro: The HDF5 Cloud-Optimized Read-Only Library
- May 14, 11:00 AM Central
- Looking at ways to efficiently access HDF5 files residing in AWS S3
- 2021 HDF5 User Group Call for Papers
- Format: "Paper and Presentation" or "Presentation"
- Deadline for abstracts: 1 June 2021
- Event: 12-15 October 2021
- H5Coro: The HDF5 Cloud-Optimized Read-Only Library
- Forum
- Zlib-ng works with HDF5
- New
zlibimplementation appears to work w/ HDF5 - Much better performance according to the post
- Maybe support as a dynamic plugin?
- New
- Reference Manual in Doxygen
- Current version
- Interesting discussion around docs-like-code and maintenance
- There are different kinds of documentation
- Tasks, concepts, reference (DITAA)
- Tutorials, how-to guides, explanation, reference (Divio)
H5Idec_refhangs
- Showing an example of MPI ranks doing independent writes to different datasets w/ MPI-IO VFD
- HDF5: infinite loop closing library (v1.10.6)
- We covered file close degrees in clinic no. 10 on
- HDFql 2.3.0 Release (with EXCEL import/export support!)
- Bugfix for Java
ArrayIndexOutOfBoundsExceptionwhen registering a zero element byte array - Fixed in HDFql 2.4.0
- Bugfix for Java
- Zlib-ng works with HDF5
Tips, tricks, & insights
- Fixed- vs. variable-length string performance cage match
- Trigger: Writing and reading variable length std::string in compound data type
- Contributed by Steven (Canada Dry) Varga
Clinic 2021-05-04
Your Questions
???
Last week's highlights
- Announcements
- Recording of last week's webinar
- H5Coro: The HDF5 Cloud-Optimized Read-Only Library
- May 14, 11:00 AM Central
- Looking at ways to efficiently access HDF5 files residing in AWS S3
- 2021 HDF5 User Group Call for Papers
- Format: "Paper and Presentation" or "Presentation"
- Deadline for abstracts: 1 June 2021
- Event: 12-15 October 2021
- Forum
Tips, tricks, & insights
- What is
H5S_ALLall about?
{ __label__ fail_update, fail_fspace, fail_dset, fail_file; hid_t file, dset, fspace; unsigned mode = H5F_ACC_RDWR; char file_name[] = "d1.h5"; char dset_name[] = "σύνολο/δεδομένων"; int new_elts[6][2] = {{-1, 1}, {-2, 2}, {-3, 3}, {-4, 4}, {-5, 5}, {-6, 6}}; if ((file = H5Fopen(file_name, mode, H5P_DEFAULT)) == H5I_INVALID_HID) { ret_val = EXIT_FAILURE; goto fail_file; } if ((dset = H5Dopen2(file, dset_name, H5P_DEFAULT)) == H5I_INVALID_HID) { ret_val = EXIT_FAILURE; goto fail_dset; } // get the dataset's dataspace if ((fspace = H5Dget_space(dset)) == H5I_INVALID_HID) { ret_val = EXIT_FAILURE; goto fail_fspace; } // select the first 5 elements in odd positions if (H5Sselect_hyperslab(fspace, H5S_SELECT_SET, (hsize_t[]){1}, (hsize_t[]){2}, (hsize_t[]){5}, NULL) < 0) { ret_val = EXIT_FAILURE; goto fail_update; } // (implicitly) select and write the first 5 elements of the second // column of NEW_ELTS if (H5Dwrite(dset, H5T_NATIVE_INT, H5S_ALL, fspace, H5P_DEFAULT, new_elts) < 0) ret_val = EXIT_FAILURE; fail_update: H5Sclose(fspace); fail_fspace: H5Dclose(dset); fail_dset: H5Fclose(file); fail_file:; }
Coming soon
- Fixed- vs. variable-length string performance cage match
- Contributed by Steven (Canada Dry) Varga
- You don't want to miss that one!
- What happens to open HDF5 handles/IDs when your program ends?
- Suggested by Quincey Koziol (LBNL)
- We'll take it in pieces
- Current behavior
- How async I/O changes that picture
- Other topics of interest?
Let us know!
Clinic 2021-04-27
Your questions
- Question 1
Last week you mentioned that one might use the Fortran version of the HDF5 library from C/C++ when working with column-major data. Could you say more about this? Is the difference simply how the arguments to the library functions are interpreted (e.g
H5Screate,H5Sselect_hyperslab) are interpreted, or is it possible to discern from the file itself whether the data is column-major or row-major?
Last week's highlights
- Announcements
- ECP BOF Day slides are available
- Part 2 of HDF5 Application Tuning
- April 30, 11:00 AM Central
- This time it's about HDF5 library performance variability
- H5Coro: The HDF5 Cloud-Optimized Read-Only Library
- May 14, 11:00 AM Central
- Looking at ways to efficiently access HDF5 files residing in AWS S3
- 2021 HDF5 User Group Call for Papers
- Format: "Paper and Presentation" or "Presentation"
- Deadline for abstracts: 1 June 2021
- Event: 12-15 October 2021
- Forum
- Doxygen-based RM merged to
develop
- Parallel HDF5 write with irregular size in one dimension
- Rob Latham confirmed an issue in Darshan's HDF5 module
- How to use H5ocopy to copy a group with hard links
- I don't understand it: are we copying groups or links (or both)?
- Doxygen-based RM merged to
Tips, tricks, & insights
- The
h5stattool
Usage: h5stat [OPTIONS] file OPTIONS -h, --help Print a usage message and exit -V, --version Print version number and exit -f, --file Print file information -F, --filemetadata Print file space information for file's metadata -g, --group Print group information -l N, --links=N Set the threshold for the # of links when printing information for small groups. N is an integer greater than 0. The default threshold is 10. -G, --groupmetadata Print file space information for groups' metadata -d, --dset Print dataset information -m N, --dims=N Set the threshold for the dimension sizes when printing information for small datasets. N is an integer greater than 0. The default threshold is 10. -D, --dsetmetadata Print file space information for datasets' metadata -T, --dtypemetadata Print datasets' datatype information -A, --attribute Print attribute information -a N, --numattrs=N Set the threshold for the # of attributes when printing information for small # of attributes. N is an integer greater than 0. The default threshold is 10. -s, --freespace Print free space information -S, --summary Print summary of file space information --enable-error-stack Prints messages from the HDF5 error stack as they occur --s3-cred=<cred> Access file on S3, using provided credential <cred> :: (region,id,key) If <cred> == "(,,)", no authentication is used. --hdfs-attrs=<attrs> Access a file on HDFS with given configuration attributes. <attrs> :: (<namenode name>,<namenode port>, <kerberos cache path>,<username>, <buffer size>) If an attribute is empty, a default value will be used.Let's see this in action:
File information # of unique groups: 718 # of unique datasets: 351 # of unique named datatypes: 4 # of unique links: 353 # of unique other: 0 Max. # of links to object: 701 Max. # of objects in group: 350 File space information for file metadata (in bytes): Superblock: 48 Superblock extension: 0 User block: 0 Object headers: (total/unused) Groups: 156725/16817 Datasets(exclude compact data): 129918/538 Datatypes: 1474/133 Groups: B-tree/List: 21656 Heap: 33772 Attributes: B-tree/List: 0 Heap: 0 Chunked datasets: Index: 138 Datasets: Heap: 0 Shared Messages: Header: 0 B-tree/List: 0 Heap: 0 Free-space managers: Header: 0 Amount of free space: 0 Small groups (with 0 to 9 links): # of groups with 0 link(s): 1 # of groups with 1 link(s): 710 # of groups with 2 link(s): 1 # of groups with 3 link(s): 2 # of groups with 4 link(s): 1 # of groups with 5 link(s): 1 Total # of small groups: 716 Group bins: # of groups with 0 link: 1 # of groups with 1 - 9 links: 715 # of groups with 100 - 999 links: 2 Total # of groups: 718 Dataset dimension information: Max. rank of datasets: 1 Dataset ranks: # of dataset with rank 1: 351 1-D Dataset information: Max. dimension size of 1-D datasets: 736548 Small 1-D datasets (with dimension sizes 0 to 9): # of datasets with dimension sizes 1: 1 Total # of small datasets: 1 1-D Dataset dimension bins: # of datasets with dimension size 1 - 9: 1 # of datasets with dimension size 100000 - 999999: 350 Total # of datasets: 351 Dataset storage information: Total raw data size: 9330522 Total external raw data size: 0 Dataset layout information: Dataset layout counts[COMPACT]: 0 Dataset layout counts[CONTIG]: 0 Dataset layout counts[CHUNKED]: 351 Dataset layout counts[VIRTUAL]: 0 Number of external files : 0 Dataset filters information: Number of datasets with: NO filter: 1 GZIP filter: 0 SHUFFLE filter: 350 FLETCHER32 filter: 0 SZIP filter: 0 NBIT filter: 0 SCALEOFFSET filter: 0 USER-DEFINED filter: 350 Dataset datatype information: # of unique datatypes used by datasets: 4 Dataset datatype #0: Count (total/named) = (1/1) Size (desc./elmt) = (60/64) Dataset datatype #1: Count (total/named) = (347/0) Size (desc./elmt) = (14/1) Dataset datatype #2: Count (total/named) = (2/0) Size (desc./elmt) = (14/2) Dataset datatype #3: Count (total/named) = (1/1) Size (desc./elmt) = (79/12) Total dataset datatype count: 351 Small # of attributes (objects with 1 to 10 attributes): # of objects with 1 attributes: 1 # of objects with 2 attributes: 551 # of objects with 3 attributes: 147 # of objects with 4 attributes: 2 # of objects with 5 attributes: 4 # of objects with 6 attributes: 1 Total # of objects with small # of attributes: 706 Attribute bins: # of objects with 1 - 9 attributes: 706 Total # of objects with attributes: 706 Max. # of attributes to objects: 6 Free-space persist: FALSE Free-space section threshold: 1 bytes Small size free-space sections (< 10 bytes): Total # of small size sections: 0 Free-space section bins: Total # of sections: 0 File space management strategy: H5F_FSPACE_STRATEGY_FSM_AGGR File space page size: 4096 bytes Summary of file space information: File metadata: 343731 bytes Raw data: 9330522 bytes Amount/Percent of tracked free space: 0 bytes/0.0% Unaccounted space: 5582 bytes Total space: 9679835 bytes
Clinic 2021-04-20
Your questions
Last week's highlights
- Announcements
- MeetingC++ online Tool Fair TODAY
- Part 2 of HDF5 Application Tuning
- This time it's about HDF5 library performance variability
- H5Coro: The HDF5 Cloud-Optimized Read-Only Library
- Looking at ways to efficiently access HDF5 files residing in AWS S3
- 2021 HDF5 User Group Call for Papers
- Format: "Paper and Presentation" or "Presentation"
- Deadline for abstracts: 1 June 2021
- Event: 12-15 October 2021
- Forum
- Parallel HDF5 write with irregular size in one dimension
- Potential issue in Darshan's HDF5 module
- Independent datasets for MPI processes. Progress?
- Discussing data life cycle and design options
- Competing requirements
- For better or worse, there are often many ways to do one thing in HDF5
- Robustness
- Discussing data life cycle and design options
- Increases the use of the system RAM until it reaches RAM saturation
- Long running application w/ no apparent handle leakage
- Valgrind
- Calling fortran HDF5 function h5dwritef from C++
- I'm out of my depth when it comes to Fortran :-(
- Parallel HDF5 write with irregular size in one dimension
Tips, tricks, & insights
- Do I need a degree to use
H5Pset_fclose_degree?
- Identifiers are transient runtime handles to manage HDF5 things
- Everything begins with a file handle, but how does it end?
- Files can be re-opened
- Other files can be mounted in HDF5 groups
- Traversal of external links may trigger the opening of other files and
objects, but see
H5Pset_elink_file_cache_size
- What happens if a file is closed before other (non-file) handles?
H5F_CLOSE_WEAK- File is closed if last open handle
- Invalidate file handle and delay file close until remaining objects are closed
H5F_CLOSE_SEMI- File is closed if last open handle
H5Fclosegenerates error if open handles remain
H5F_CLOSE_STRONG- File is closed, closing any remaining handles if necessary.
H5F_CLOSE_DEFAULT- VFD decides,
H5F_CLOSE_WEAKfor most VFDs. Notable exception: MPI-IO -H5F_CLOSE_SEMI
Clinic 2021-04-06
Your questions
- Question 1
We have observed that reading a dataset with variable-length ASCII strings and setting the read mem. type to
H5T_C_S1 (size=H5T_VARIABLE / cset=H5T_CSET_UTF8), produces an error with“H5T.c line 4893 in H5T__path_find_real(): no appropriate function for conversion path”. However, if we read first another dataset of the same file that contains UTF8 strings and then the same dataset with ASCII strings, no errors are returned whatsoever and the content seems to be retrieved. Is this an expected behaviour, or are we missing something?- As a side note, the same situation can be replicated by setting the cset to
H5T_CSET_ASCIIand opening first the ASCII-based dataset before the UTF8-dataset, or any other combination, as long as the first call succeeded (e.g., opening the ASCII dataset withcset=H5T_CSET_ASCII, then opening the same ASCII dataset withcset=H5T_CSET_UTF8also seems to work). - Tested using HDF5 v1.10.7, v1.12.0, and manually compiling the most recent
commit on the official GitHub repository. The code was compiled with GCC
9.3.0 + HPE-MPI v2.22, but no MPI file access property was given (i.e., using
H5P_DEFAULTto avoid MPI-IO). - Further information: https://github.com/HDFGroup/hdf5/issues/544
- As a side note, the same situation can be replicated by setting the cset to
Last week's highlights
- Announcements
- Alpha 4 release of HDF5.NET
- Based on the HDF5 file format spec. & no HDF5 library dependence!
- 2021 HDF5 User Group Call for Papers
- Format: "Paper and Presentation" or "Presentation"
- Deadline for abstracts: 1 June 2021
- Event: 12-15 October 2021
- Alpha 4 release of HDF5.NET
- Forum
- How can attributes of an existing object be modified?
- There are several different "namespaces" in HDF5
- Examples:
- Global (=file-level) path names
- Per object attribute names
- Per compound type field names
- Etc.
- Some have constraints such as reserved characters, character encoding, length, etc.
- Most importantly, they are disjoint and don't mix
- Disambiguation would be too costly, if not impossible
- HDF5DotNet library
- There's perhaps a place for wrappers of the HDF5 C-API and and independent .NET native (=full-managed) solution (e.g., HDF5.NET)
- SWIG (Simplified Wrapper and Interface Generator) has come a long way
- Should that be the path forward for HDF.PInvoke
- We need greater automation and (.NET) platform independence
- Focus on testing
- Any thoughts/comments?
- Parallel HDF5 write with irregular size in one dimension
- Posted an example that shows how different ranks can write varying amounts of data to a chunked dataset in parallel. Some ranks don't write any data. The chunk size is chosen arbitrarily.
- How can attributes of an existing object be modified?
Tips & tricks
- The "mystery" of the HDF5 file format
- The specification published here can seem overwhelming. Part of the problem is that you are seeing at least three versions layered on top of each other.
- The first (?) release was a lot simpler, and has all the core ideas
- Once you've digested that, you are ready for the other releases and consider writing your own (de-)serializer
- Don't get carried away: only a tiny fraction of the HDF5 library's code deals w/ serialization
Clinic 2021-03-30
Canceled because of ECP event.
Clinic 2021-03-23
Your questions
???
Last week's highlights
- Announcements
- 2021 HDF5 User Group Call for Papers
- Format: "Paper and Presentation" or "Presentation"
- Deadline for abstracts: 1 June 2021
- Event: 12-15 October 2021
- HDF4 is on GitHub
- Don't forget to register for the Hermes webinar on Friday (03/26)
- 2021 HDF5 User Group Call for Papers
- Forum
- How to convert XML to HDF5
- There is no canonical conversion path, even if you have an XML schema
- XML is simpler because elements are strictly nested
- XML can be trickier because of element repetition and the non-obligatory nature of certain elements or attributes
- Start w/ a scripting language that has XML (parsing) and HDF5 modules
- Jannson works well if you prefer C
- Consider XSLT to simplify first
- There is no canonical conversion path, even if you have an XML schema
- HDF5DotNet library
- It's been out of maintenance for many years
- Alternatives: HDF.PInvoke (Windows only) and HDF.PInvoke.1.10 (.NET
Standard)
- Both are based on HDF5 1.10.x
- Note: We (The HDF Group) are neither C# nor .NET experts.
PInvokeis about the level of abstraction we can handle. We count on and rely on knowledgeable community members for advice and contributions. - There are many interesting community projects, for example, HDF5.NET:
- Based on the HDF5 file format spec. & no HDF5 library dependence!
- Parallel HDF5 write with irregular size in one dimension
- Many of our examples s..k, and we have to do a lot better
- Maybe we created them this way to generate more questions? :-/
- HDF5 dataspaces are logical, chunks are physical
- Write a (logically) correct program first and then optimize performance!
- Many of our examples s..k, and we have to do a lot better
- How to convert XML to HDF5
Tips & tricks
- Large (> 64 KiB) HDF5 attributes
import h5py, numpy as np with h5py.File('my.h5', 'w', libver='latest') as file: file.attrs['random[1024]'] = np.random.random(1024) file.attrs['random[1048576]'] = np.random.random(1024*1024)
The
h5dumpoutput looks like this:gerd@guix ~/scratch/run$ h5dump -pBH my.h5 HDF5 "my.h5" { SUPER_BLOCK { SUPERBLOCK_VERSION 3 FREELIST_VERSION 0 SYMBOLTABLE_VERSION 0 OBJECTHEADER_VERSION 0 OFFSET_SIZE 8 LENGTH_SIZE 8 BTREE_RANK 16 BTREE_LEAF 4 ISTORE_K 32 FILE_SPACE_STRATEGY H5F_FSPACE_STRATEGY_FSM_AGGR FREE_SPACE_PERSIST FALSE FREE_SPACE_SECTION_THRESHOLD 1 FILE_SPACE_PAGE_SIZE 4096 USER_BLOCK { USERBLOCK_SIZE 0 } } GROUP "/" { ATTRIBUTE "random[1024]" { DATATYPE H5T_IEEE_F64LE DATASPACE SIMPLE { ( 1024 ) / ( 1024 ) } } ATTRIBUTE "random[1048576]" { DATATYPE H5T_IEEE_F64LE DATASPACE SIMPLE { ( 1048576 ) / ( 1048576 ) } } } }The
libver='latest'keyword is critical. Running without produces this error:gerd@guix ~/scratch/run$ python3 large_attribute.py Traceback (most recent call last): File "large_attribute.py", line 6, in <module> file.attrs['random[1048576]'] = np.random.random(1024*1024) File "h5py/_objects.pyx", line 54, in h5py._objects.with_phil.wrapper File "h5py/_objects.pyx", line 55, in h5py._objects.with_phil.wrapper File "/home/gerd/.guix-profile/lib/python3.8/site-packages/h5py/_hl/attrs.py", line 100, in __setitem__ self.create(name, data=value) File "/home/gerd/.guix-profile/lib/python3.8/site-packages/h5py/_hl/attrs.py", line 201, in create attr = h5a.create(self._id, self._e(tempname), htype, space) File "h5py/_objects.pyx", line 54, in h5py._objects.with_phil.wrapper File "h5py/_objects.pyx", line 55, in h5py._objects.with_phil.wrapper File "h5py/h5a.pyx", line 47, in h5py.h5a.create RuntimeError: Unable to create attribute (object header message is too large)libver=('v108', 'v108')also works. (v108corresponds to HDF5 1.8.x).
Clinic 2021-03-16
Your questions
???
Last week's highlights
- Announcements
- Webinar Announcement: Hermes – A Distributed Buffering System for Heterogeneous Storage Hierarchies
- HSDS version 0.6.3 released
- Grab the latest image on Docker Hub
hdfgroup/hsds:v0.6.3!
- Grab the latest image on Docker Hub
- Forum
- Multithreaded writing to a single file in C++
- Beware of non-thread-safe wrappers or language bindings!
- Compiling the C library with
--enable-threadsafeis only the first step
- Reference Manual in Doxygen
H5Iget_namecall is very slow for HDF5 file > 5 GB
H5Iget_nameconstructs an HDF5 path name given an object identifier- Use Case: You are in a corner of an application where all you've got is a handle (identifier) and you would like to render something meaningful to humans.
- It's not so much the file size but the number and arrangement of objects that
makes
H5Iget_nameslow- See the
h5statoutput the user provided!
- See the
- What contributes to
H5Iget_namebeing slow?- The path names are not stored in an HDF5 file (except in symbolic links…) and are created on-demand
- In general, HDF5 arrangements are not trees, not even directed graphs, but
directed multi-graphs
- A node can be the target of multiple edges (including from the same source node)
- Certain nodes (groups) can be source and target of an edge
- *Take-Home-Message:*Unless you are certain that your HDF5 arrangement is a
tree, you are skating on thin ice with path names!
- Trying to uniquely identify objects via path name is asking for trouble
- Use addresses + file IDs (pre-HDF 1.12) or tokens (HDF 1.12+) for that!
- Trying to uniquely identify objects via path name is asking for trouble
- Quincey points out that
- The library caches metadata that can accelerate
H5Iget_name But there are other complications
- For example, you can have "anonymous" objects (objects that haven't
been linked to groups in the file. i.e., no path yet)
- Another source of trouble are objects that have been unlinked
- The library caches metadata that can accelerate
- Multithreaded writing to a single file in C++
Tips & tricks
- How to open an HDF5 in append mode?
To be clear, there is no
H5F*call that behaves like an append call. But we can mimic one as follows:Credits: Werner Benger
1: 2: hid = H5Fcreate(filename, H5F_ACC_EXCL|H5F_ACC_SWMR_WRITE, fcpl_id, fapl_id); 3: if (hid < 0) 4: { 5: hid = H5Fopen(filename, H5F_ACC_RDWR|H5F_ACC_SWMR_WRITE, fapl_id); 6: } 7: 8: if (hid < 0) 9: // something's going on... 10:
- If the file exists
H5Fcreatewill fail andH5FopenwithH5F_ACC_RDWRwill kick in.- If the file is not an HDF5 file, both will fail.
- If the file does not exist,
H5Fcreatewill do its job.
- If the file exists
Clinic 2021-03-09
Your questions (as of 9:00 a.m. Central Time)
- Question 1
Is there a limit on array size if I save an array as an attribute of a dataset?
In terms of the performance, is there any consequence if I save a large amount of data into an attribute?
- Size limit
- No, not in newer versions (1.8.x+) of HDF5. See What limits are there in HDF5?
- Make sure that downstream applications can handle such attributes (i.e., use HDF5 1.8.x or later)
- Remember to tell the library that you want to use the 1.8 or later file
format via
H5Fset_libverbounds(e.g., setlowtoH5F_LIBVER_V18) - Also keep an eye on
H5Pset_attr_phase_change(Consider settingmax_compactto 0.)
- Performance
- It depends. (…on what you mean by performance)
- Attributes have a different function (from datasets) in HDF5
- They "decorate" other objects - application metadata
- Their values are treated as atomic units, i.e., you will always write and
read the entire "large" value.
- In other words, you lose partial I/O
- Several layouts available for datasets are not supported with attributes
- No compression
- Attributes have a different function (from datasets) in HDF5
- Question 2
Question regarding hdf5 I/O performance, compare saving data into a large array in one dataset Vs saving data into several smaller arrays and in several dataset. Any consequence in terms of the performance? Will there be any sweet spot for best performance? Or any tricks to make it reading/writing faster? I know parallel I/O but parallel I/O would need hardware support which is not always available. So the question is about the tricks to speed up I/O without parallel I/O.
- One large dataset vs. many small datasets, which is faster?
- It depends.
- How do you access the data?
- Do you always write/read the entire array in the order it was written?
- Is it WORM (write once read many)?
- How and how frequently does it change?
- How compressible is the data?
- Do you need to store data at all? E.g., HDF5-UDF
- What is performance for you and how do you measure it?
- What percentage of total runtime does your application spend doing I/O?
- What scalability behavior do you expect?
- Assuming throughput is the measure, create a baseline for your target
system, for example, via FIO or IOR
- Your goal is to saturate the I/O subsystem
- Is this a dedicated system?
- Which other systems do you need to support? Are you the only user? What's the future?
- What's the budget?
- How do you access the data?
Last week's highlights
- Announcements
- h5py 3.2 release
- Supports the HDF5 S3 (read-only) VFD thanks to Satrajit Ghosh
- Need to build h5py from source
- Minimum Python version 3.7
- Interesting bugfix: Fix reading data with a datatype of variable-length arrays of fixed length strings
- Supports the HDF5 S3 (read-only) VFD thanks to Satrajit Ghosh
- HDF5 Community BOF at ECP Community BOF Days
- March 30, 3:00 p.m. (Eastern)
- Registration
- h5py 3.2 release
- Forum
- Get Object Header size
The user created a compound type with 100s of fields and eventually saw this error:
H5Oalloc.c line 1312 in H5O__alloc(): object header message is too large
- This issue was first raised (Jira-ticket HDFFV-1089 date) on Jun 08, 2009
- Root cause: the size of header message data is represented in a 2 byte
unsigned integer (see section IV.A.1.a and IV.A.1.b of the HDF5 file format spec.)
- Ergo, header messages, currently, cannot be larger than 64 KB.
- Datatype information is stored in a header message (see section IV.A.2.d)
- This can be fixed with a file format update, but it's fallen through the cracks for over 10 years
- The customer is always right, but who needs 100s of fields in a compound type?
- Use Case: You have a large record type and you always (or most of the time) read and write all fields together.
- Outside this narrow use case you are bound to lose a lot of performance and flexibility
- You are Leaving the
American SectorMainstream: not too many tools will be able to handle your data - Better approach: divide-and-conquer, i.e., go w/ a group of compounds or individual columns
- Using HDF5 in Qt Creator
- Linker can't find
H5::FileAccPropList()andH5::FileCreatPropList() - Works fine in release mode, but not in debug mode
- AFAIK, we don't distribute debug libraries in binary form. Still doesn't explain why the user couldn't use the release binaries in a debug build, unless QT Creator is extra pedantic?
- Linker can't find
- Reference Manual in Doxygen
H5Iget_namecall is very slow for HDF5 file > 5 GB
H5Iget_nnameconstructs an HDF5 path name given an object identifier- Use Case: You are in a corner of an application where all you've got is a handle (identifier) and you would like to render something meaningful to humans.
- It's not so much the file size but the number and arrangement of objects that
makes
H5Iget_nameslow- See the
h5statoutput the user provided!
- See the
- What contributes to
H5Iget_namebeing slow?- The path names are not stored in an HDF5 file (except in symbolic links…) and are created on-demand
- In general, HDF5 arrangements are not trees, not even directed graphs, but
directed multi-graphs
- A node can be the target of multiple edges (including from the same source node)
- Certain nodes (groups) can be source and target of an edge
- *Take-Home-Message:*Unless you are certain that your HDF5 arrangement is a
tree, you are skating on thin ice with path names!
- Trying to uniquely identify objects via path name is asking for trouble
- Use addresses + file IDs (pre-HDF 1.12) or tokens (HDF 1.12+) for that!
- Trying to uniquely identify objects via path name is asking for trouble
- Get Object Header size
Clinic 2021-03-02
Your questions
- h5rnd
- Question: How are generated HDF5 objects named? An integer name, or can a
randomized string be used?
h5rndGenerates a pool of random strings as link names- Uniform length distribution between 5 and 30 over
[a-z][A-Z]
- Question: Does it create multi-dimensional datasets with a rich set of
HDF5 datatypes? Compound datatypes, perhaps?
- Currently, it creates 1,000 element 1D FP64 datasets (w/ attribute)
- RE: types - anything is possible. Budget?
- Question: Are named datatypes generated? If not, are these reasonable
types of extensions for
h5rnd?- Not currently, but anything is possible
- Question: How are generated HDF5 objects named? An integer name, or can a
randomized string be used?
- Other questions?
- Question: How do these extensions fit with the general intent and
extensibility of
h5rnd?- It was written as an illustration
- Uses an older version of H5CPP
- Labeling could be improved
- Dataset generation under development
- Some enhancements in a future version
- Question: How do these extensions fit with the general intent and
extensibility of
Last week's highlights
- Forum
- External link access in parallel HDF5 1.12.0
- Can't access externally linked datasets in parallel; fine in 1.10.x and in serial
- It appears that someone encountered a known bug in the field
- Dev. claim it's fixed in
develop, waiting for confirmation from the user
H5I_dec_refhangs
H5Idec_refis one of those functions that needs to be used w/ extra care- Using
mpi4pyandh5py - User provided an MWE (in Python) and, honestly, there is limited help we can
offer (as we are neither
mpi4pynorh5pyexperts) - A C or C++ MWE might be the better starting point
h5diffexits with 1 but doesn’t print differences
- Case of out-of-date/poor documentation
h5diffis perhaps the most complex tool (multi-graph comparison + what does'='mean?)- Writing code is the easy part
- We need to do better
- Independent datasets for MPI processes. Progress?
- Need some clarification on the problem formulation
- Current status (w/ MPI) MD-modifying ops. must be collective
- On the horizon: asynchronous operations (ASYNC VOL)
- Writing to virtual datasets
- Apparently broken when a datatype conversion (truncation!) is involved
- External link access in parallel HDF5 1.12.0
Clinic 2021-02-23
Your questions
- How to use
H5Ocopyin C++ code?
-
sandhya.v250 (Feb 19)
Hello Team, I want to copy few groups from one hdf5 file to hdf5 another file which is not yet created and this should be done inside the C++ code..can you please tell me how can I use this inside this tool
The function in question (there is also a tool called
h5copy):herr_t H5Ocopy ( hid_t src_loc_id, const char* src_name, hid_t dst_loc_id, const char* dst_name, hid_t ocpypl_id, hid_t lcpl_id );
- The emphasis appears to be on C++
- You can do this in C. It's just more boilerplate.
- Whenever I need something C++, I turn to my colleague Steven Varga (= Mr. H5CPP)
- He also created a nice random HDF5 file generator/tester (= 'Prüfer' in German)
-
- Steven's solution (excerpt)
The full example can be downloaded from here.
Basic idea: Visit all objects in the source via
H5Ovisitand invokeH5Ocopyin the callback.1: 2: #include "argparse.h" 3: #include <h5cpp/all> 4: #include <string> 5: 6: herr_t ocpy_callback(hid_t src, const char *name, const H5O_info_t *info, 7: void *dst_) { 8: hid_t* dst = static_cast<hid_t*>(dst_); 9: int err = 0; 10: switch( info->type ){ 11: case H5O_TYPE_GROUP: 12: if(H5Lexists( *dst, name, H5P_DEFAULT) >= 0) 13: err = H5Ocopy(src, name, *dst, name, H5P_DEFAULT, H5P_DEFAULT); 14: break; 15: case H5O_TYPE_DATASET: 16: err = H5Ocopy(src, name, *dst, name, H5P_DEFAULT, H5P_DEFAULT); 17: break; 18: default: /*H5O_TYPE_NAMED_DATATYPE, H5O_TYPE_NTYPES, H5O_TYPE_UNKNOWN */ 19: ; // nop to keep compiler happy 20: } 21: return 0; 22: } 23: 24: int main(int argc, char **argv) 25: { 26: argparse::ArgumentParser arg("ocpy", "0.0.1"); 27: arg.add_argument("-i", "--input") 28: .required().help("path to input hdf5 file"); 29: arg.add_argument("-s", "--source") 30: .default_value(std::string("/")) 31: .help("path to group within hdf5 container"); 32: arg.add_argument("-o", "--output").required() 33: .help("the new hdf5 will be created/or opened rw"); 34: arg.add_argument("-d", "--destination") 35: .default_value(std::string("/")) 36: .help("target group"); 37: 38: std::string input, output, source, destination; 39: try { 40: arg.parse_args(argc, argv); 41: input = arg.get<std::string>("--input"); 42: output = arg.get<std::string>("--output"); 43: source = arg.get<std::string>("--source"); 44: destination = arg.get<std::string>("--destination"); 45: 46: h5::fd_t fd_i = h5::open(input, H5F_ACC_RDONLY); 47: h5::fd_t fd_o = h5::create(output, H5F_ACC_TRUNC); 48: h5::gr_t dgr{H5I_UNINIT}, sgr = h5::gr_t{H5Gopen(fd_i, source.data(), 49: H5P_DEFAULT)}; 50: h5::mute(); 51: if( destination != "/" ){ 52: char * gname = destination.data(); 53: dgr = H5Lexists(fd_o, gname, H5P_DEFAULT) >= 0 ? 54: h5::gr_t{H5Gcreate(fd_o, gname, H5P_DEFAULT, H5P_DEFAULT, 55: H5P_DEFAULT)} 56: : h5::gr_t{H5Gopen(fd_i, gname, H5P_DEFAULT)}; 57: H5Ovisit(sgr, H5_INDEX_CRT_ORDER, H5_ITER_NATIVE, ocpy_callback, &dgr ); 58: } else 59: H5Ovisit(sgr, H5_INDEX_CRT_ORDER, H5_ITER_NATIVE, ocpy_callback, &fd_o); 60: h5::unmute(); 61: } catch ( const h5::error::any& e ) { 62: std::cerr << e.what() << std::endl; 63: std::cout << arg; 64: } 65: return 0; 66: } 67:
- Parting thoughts
- This is can be tricky business depending on how selective you want to be
H5Ovisitvisits objects and does not account for dangling links, etc.H5Ocopy's behavior is highly customizable. Check the options & play w/h5copyto see the effect!
- More Questions
- Question 1
I have an unrelated question. I have 7,000 HDF5 files, each 500 MB long. When I use them, should I open them selectively, when I need them, or is it advantageous to make one big file, or to open virtual files? I am interested in the speed of the different approaches.
- 40 GbE connectivity
- 10 contiguously laid out Datasets per file => ~50 MB per dataset
- Always reading full datasets
- Considerations:
- If you have the RAM and use all data in an "epoch" just read whole files and use HDF5 file images for "in-memory I/O."
- You could maintain an index file I which contains external links (one for each of the 7,000 files), and a dataset which for each external file and dataset contains the offset of the dataset in the file. You would keep I (small!) in memory and, for each dataset request, read the ~50MB directly w/o the HDF5 library. This assumes that no datatype conversion is necessary and you have no trouble interpreting the bytes.
- A variation of the previous approach would be for the stub-file to contain HDF5 virtual datasets, datasets stitched together from other datasets. This would we a good option, if you wanted to simplify your application code and make everything appear as a single large HDF5 file. It'd be important though to have that (small) stub-file on the clients in memory to not incur a high latency penalty.
- Both approaches can be easily parallelized, assuming read-only access. If there are writers involved, it's still doable, but additional considerations apply.
Another question: what is the recommended way to combine Python with C++ with C++ reading in and working on large hdf5 files that require a lot of speed.
- To be honest, we ran out of time and I (GH) didn't fully grasp the question.
- Steven said something about Julia
- Henric uses Boost Python. What about Cython?
What's the access pattern?
Let's continue the discussion on the forum or come back next week!
- Question 1
Last week's highlights
- Forum
- Hyperslab selection bug confirmed
- Write data to variable length string attribute bug is a feature
;-) - Possible bug in HDFView
- The MWE checks out and it looks more and more like a bug in HDFView
Appendix
- The
h5copycommand line tool
gerd@guix ~$ h5copy usage: h5copy [OPTIONS] [OBJECTS...] OBJECTS -i, --input input file name -o, --output output file name -s, --source source object name -d, --destination destination object name OPTIONS -h, --help Print a usage message and exit -p, --parents No error if existing, make parent groups as needed -v, --verbose Print information about OBJECTS and OPTIONS -V, --version Print version number and exit --enable-error-stack Prints messages from the HDF5 error stack as they occur. -f, --flag Flag type Flag type is one of the following strings: shallow Copy only immediate members for groups soft Expand soft links into new objects ext Expand external links into new objects ref Copy references and any referenced objects, i.e., objects that the references point to. Referenced objects are copied in addition to the objects specified on the command line and reference datasets are populated with correct reference values. Copies of referenced datasets outside the copy range specified on the command line will normally have a different name from the original. (Default:Without this option, reference value(s) in any reference datasets are set to NULL and referenced objects are not copied unless they are otherwise within the copy range specified on the command line.) noattr Copy object without copying attributes allflags Switches all flags from the default to the non-default setting These flag types correspond to the following API symbols H5O_COPY_SHALLOW_HIERARCHY_FLAG H5O_COPY_EXPAND_SOFT_LINK_FLAG H5O_COPY_EXPAND_EXT_LINK_FLAG H5O_COPY_EXPAND_REFERENCE_FLAG H5O_COPY_WITHOUT_ATTR_FLAG H5O_COPY_ALL
Clinic 2021-02-09
THIS MEETING IS BEING RECORDED and the recording will be available on The HDF Group's YouTube channel. Remember to subscribe!
Goal(s)
This is a meeting dedicated to your questions.
In the unlikely event there aren't any
We have a few prepared topics (forum posts, announcements, etc.)
Sometimes life deals you an HDF5 file
No question is too small. We are here to learn. All of us.
Meeting Etiquette
Be social, turn on your camera (if you've got one)
Talking to black boxes isn't fun.
Raise your hand to signal a contribution (question, comment)
Mute yourself while others are speaking, be ready to participate.
Be mindful of your "airtime"
We want to cover as many of your topics as possible. Be fair to others.
Introduce yourself
- Your Name
- Your affiliation/organization/group
- One reason why you are here today
Use the shared Google doc for questions and code snippets
The link can be found in the chat window.
When the 30 min. timer runs out, this meeting is over.
Continue the discussion on the HDF Forum or come back next week!
Notes
Don't miss our next webinar about data virtualization with HDF5-UDF and how it can streamline your work
- Presented by Lucas Villa Real (IBM Research)
- Feb 12, 2021 11:00 AM in Central Time (US and Canada)
- Sign-up link
Bug-of-the-Week Award (my candidate)
- Write data to variable length string attribute by Kerim Khemraev
- Jira issue HDFFV-11215
Quick demonstration
#include "hdf5.h" #include <filesystem> #include <iostream> #include <string> #define H5FILE_NAME "Attributes.h5" #define ATTR_NAME "VarLenAttr" namespace fs = std::filesystem; int main(int argc, char *argv[]) { hid_t file, attr; auto attr_type = H5Tcopy(H5T_C_S1); H5Tset_size(attr_type, H5T_VARIABLE); H5Tset_cset(attr_type, H5T_CSET_UTF8); auto make_scalar_attr = [](auto& file, auto& attr_type) -> hid_t { auto attr_space = H5Screate(H5S_SCALAR); auto result = H5Acreate(file, ATTR_NAME, attr_type, attr_space, H5P_DEFAULT, H5P_DEFAULT); H5Sclose(attr_space); return result; }; if( !fs::exists(H5FILE_NAME) ) { // If the file doesn't exist we create it & // add a root group attribute std::cout << "Creating file...\n"; file = H5Fcreate(H5FILE_NAME, H5F_ACC_TRUNC, H5P_DEFAULT, H5P_DEFAULT); attr = make_scalar_attr(file, attr_type); } else { // File exists: we either delete the attribute and // re-create it, or we just re-write it. std::cout << "Opening file...\n"; file = H5Fopen(H5FILE_NAME, H5F_ACC_RDWR, H5P_DEFAULT); #ifndef REWRITE_ONLY H5Adelete(file, ATTR_NAME); attr = make_scalar_attr(file, attr_type); #else attr = H5Aopen(file, ATTR_NAME, H5P_DEFAULT); #endif } // Write or re-write the attribute const char* data[1] = { "Let it be λ!" }; H5Awrite(attr, attr_type, data); hsize_t size; H5Fget_filesize(file, &size); std::cout << "File size: " << size << " bytes\n"; H5Tclose(attr_type); H5Aclose(attr); H5Fclose(file); }
Documentation update
- Doxygen-based RM
- Remaining: a few
H5Pcalls andH5E - Current version: https://hdf5.io/develop/modules.html
Clinic 2021-02-16
Your questions
Last week's highlights
- Events
- Forum
- Release of HDFql 2.3.0
- Excel import and export (no Excel or OLE dependency!)
- Possible bug in HDFView
- Based on the description, it appears that external links are not properly traversed
- Waiting for MWE
- Possible hyperslab selection bug in 1.10.7
- It appears that
H5S_SELECT_ORis no longer "commutative", i.e.,H5DreadwithSelA OR SelBworks, but notSelB OR SelA - Wasn't there in 1.10.6
- It appears that
- Release of HDFql 2.3.0
Notes
- What (if any) are the ACID properties of HDF5 operations?
- Split-state
The state of an open (for RW) HDF5 file is split between RAM and persistent storage. Often the partial states will be out of sync. In the event of a "catastrophic" failure (power outage, application crash, system crash), it is impossible to predict what the partial state on disk will be.
skinparam componentStyle rectangle package "HDF5 File State" { database "Disk" { [Partial State 1] } cloud "RAM" { [Partial State 2] } }

- Non-transactional
The main reason why it is impossible to predict the outcome is that HDF5 operations are non-transactional. By 'transaction' I mean a collection of operations (and the effects of their execution) on the physical and abstract application state. In particular, there are no concepts of beginning a transaction, a commit, or a roll-back. Since they are not transactional, it is not straightforward to speak about the ACID properties of HDF5 operations.
- File system facilities
People sometimes speak about ACID properties with respect to file system operations. Although the HDF5 library relies on file system operations for the implementation of HDF5 operations, the correspondence is not as direct as might wish. For example, what appears as a single HDF5 operation to the user often includes multiple file system operations. Several file system operations have a certain property only at the level of a single operation, but not multiple operations combined.
- ACID
- Atomicity
- All changes to an HDF5 file's state must complete or fail as a
whole unit.
- Supported in HDF5? No.
- Some file systems only support single op. atomicity, if at all.
- A lot of HDF5 operations are in-place; mixed success -> impossible to recover
- Consistency
- An operation is a correct transformation of the HDF5 file's
state.
- Supported in HDF5? Yes and No
- Depends on one's definition of HDF5 file/object integrity constraints
- Assuming we are dealing with a correct program
- Special case w/ multiple processes: Single Writer Multiple Reader
- Isolation (serialization)
- Even though operations execute concurrently, it
appears to each operation, OP, that others executed either before OP or after
OP, but not both.
- Supported in HDF5? No.
- Depends on concurrency scenario and requires special configuration (e.g., MT, MPI).
- Time-of-check-time-of-use vulnerability
- Durability
- Once an operation completes successfully, it's changes to the
file's state survive failure.
- Supported in HDF5? No.
- "Split brain"
- No transaction log
- Split-state