 |
HDF5 Last Updated on 2026-02-07
The HDF5 Field Guide
|
 |
HDF5 Last Updated on 2026-02-07
The HDF5 Field Guide
|
Navigate back: Main / Technical Notes
Internal compression is one of several powerful HDF5 features that distinguish HDF5 from other binary formats and make it very attractive for storing and organizing data. Internal HDF5 compression saves storage space and I/O bandwidth and allows efficient partial access to data. Chunked storage has to be used when HDF5 compression is enabled.
Certain combinations of compression, chunked storage, and access pattern may cause I/O performance degradation if used inappropriately, but the HDF5 Library provides tuning parameters to achieve I/O performance comparable with the I/O performance on raw data that uses contiguous storage.
In this paper, we discuss the factors that should be considered when storing compressed data in HDF5 files and how to tune those parameters to optimize the I/O performance of an HDF5 application when working with compressed datasets.
One of the most powerful features of HDF5 is its ability to store and modify compressed data. The HDF5 Library comes with two pre-defined compression methods, GNU zip or gzip and szip or libaec, and has the capability to use third-party compression methods, Using Dynamically-Loadable Filters. The variety of available compression methods means users can choose the compression method that is best suited for achieving the desired balance between the CPU time needed to compress or un-compress data and storage performance.
Compressed data is stored in a data array of an HDF5 dataset using a chunked storage mechanism. When chunked storage is used, the data array is split into equally sized chunks each of which is stored separately in the file.
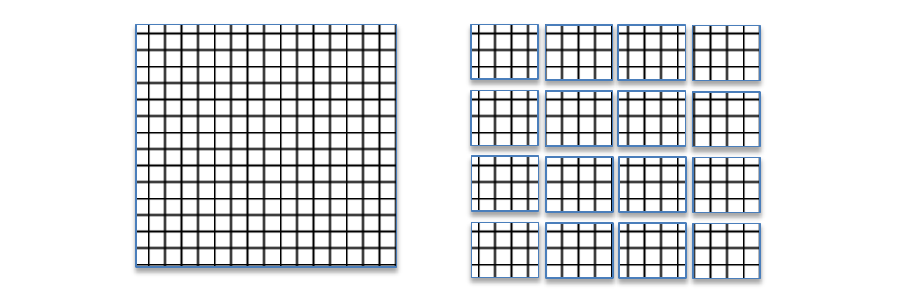
Figure 1 |
Compression is applied to each individual chunk. When an I/O operation is performed on a subset of the data array, only chunks that include data from the subset participate in I/O and need to be uncompressed or compressed.
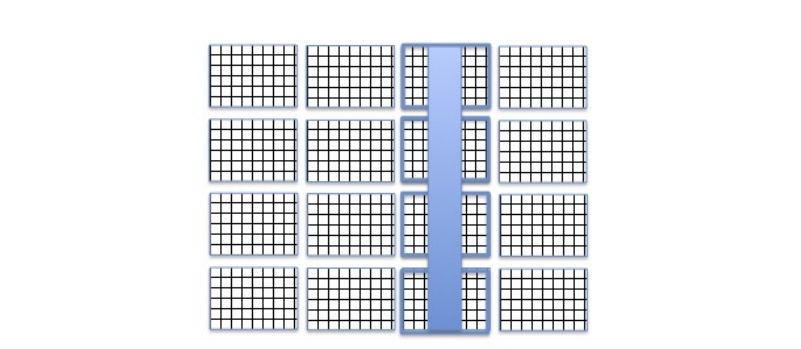
Figure 2 |
Chunked storage also enables adding more data to a dataset without rewriting the whole dataset. Figure 3 below shows more rows and columns added to a data array stored in HDF5 by writing highlighted chunks that contain new data.
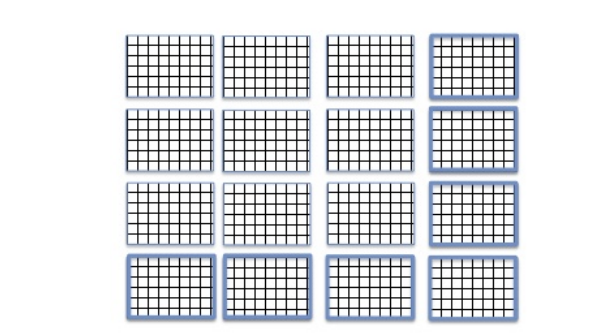
Figure 3 |
While HDF5 chunk storage and compression obviously provide great benefits in working with data, many HDF5 users have found that sometimes I/O performance is slower for compressed data than for uncompressed data. For example, as we show in this paper, there may be a huge performance difference between an application reading compressed data and reading the same data that was not compressed. For an application that writes compressed data, I/O performance may be excellent, but when data is moved to another system and read back, I/O performance drastically drops making data virtually unusable.
Many of these cases of drastically slower reading performance can be ameliorated by more careful consideration of avoiding chunking arrangements that may cause poor reading performance when creating datasets or by a few simple changes to the application reading the data. In this paper, we will discuss the factors that should be considered when storing compressed data in HDF5 files and when tuning an HDF5 application that writes or reads compressed data. We assume that the reader knows HDF5 Learning the Basics and would like to learn a set of performance tuning techniques when working with compressed data.
In our discussion, we use an HD5 file with Cross-track Infrared Sounder (CriS) data from the Suomi NPP satellite to illustrate several performance tuning techniques for HDF5 applications. The paper is organized as follows:
In the near future, we intend to make available a new CCP (Chunking and Compression Performance) tool. This tool will allow users to vary access patterns, chunk sizes, compression method, and cache settings using the tool’s command options, reducing the need to create and compile test programs such as those used in the “Case Study” section on page 7.
For more information on other things that can affect performance, see the “Things That Can Affect Performance” page in the FAQ on the website.
We will use two HDF5 files to compare I/O performance and to illustrate the issues users may encounter when working with compressed data. These HDF5 files and the application programs used to read them can be downloaded [ 7 ] by readers wishing to reproduce the performance results discussed in this paper.1
SCRIS_npp_d20140522_t0754579_e0802557_b13293_c20140522142425734814_noaa_pop.h5 is the first file we will use. It is an original data file with Cross-track Infrared Sounder (CriS) data from the Suomi NPP satellite. For brevity, we will refer to this file in this document as File.h5.
The second file is gz6_SCRIS_npp_d20140522_t0754579_e0802557_b13293__noaa_pop.h5. We will refer to this file as File_with_compression.h5. The file was created from File.h5 by the h5repack tool that applied the gzip compression to all datasets using level 6 effort. Repacking File.h5 using gzip compression reduced the storage space by 1.3 times. We will use the file to demonstrate the most common issues HDF5 users encounter when working with compressed data in HDF5.
We selected these files because they have characteristics that would be the first ones to look at when tuning I/O performance of both writing and reading HDF5 applications. First, this data file represents files generated on a big-endian system that is usually not available to general users of the data. The data provider used the HDF5 parameters to minimize storage space for data and to maximize write speed that were not necessarily the optimum parameters for the systems where the data would be read. Second, the users’ applications read data in a way that was optimized for scientific data analysis but not optimal for the HDF5 I/O performance. We will use the files to show what the users can do to improve performance of their applications, and which factors data providers should consider before creating data products.
In our case study, we used a 4-dimensional array of 32-bit big-endian floating point numbers stored in the HDF5 dataset /All_Data/CrIS-SDR_All/ES_ImaginaryLW in both files. The data array is extensible and has the current dimension sizes 60x30x9x717. When compressed with gzip compression with level 6, the compression ratio is 1.0762. We used HDF5 command line tools h5dump and h5ls and the HDF Java-based browser HDFView to find various properties of the dataset that would help us to understand performance problems and propose solutions. If the reader decides to follow the discussion using a “hands on” approach, the examples below illustrate how to use h5dump and h5ls to get the characteristics of the /All_Data/CrIS-SDR_All/ES_ImaginaryLW dataset.
The h5dump command line below will yield the results shown in Figure 4 below:
Figure 4: Output of the h5dump command that shows properties of the dataset /All_Data/CrIS-SDR_All/ES_ImaginaryLW
The h5ls command line below will yield the results shown in Figure 5 below:
Figure 5: Output of the h5ls command that shows properties of the dataset /All_Data/CrIS-SDR_All/ES_ImaginaryLW
In HDFView, right click on the dataset to choose “Show Properties” option from the drop-down menu. The properties will appear in the new window as shown in Figure 6.
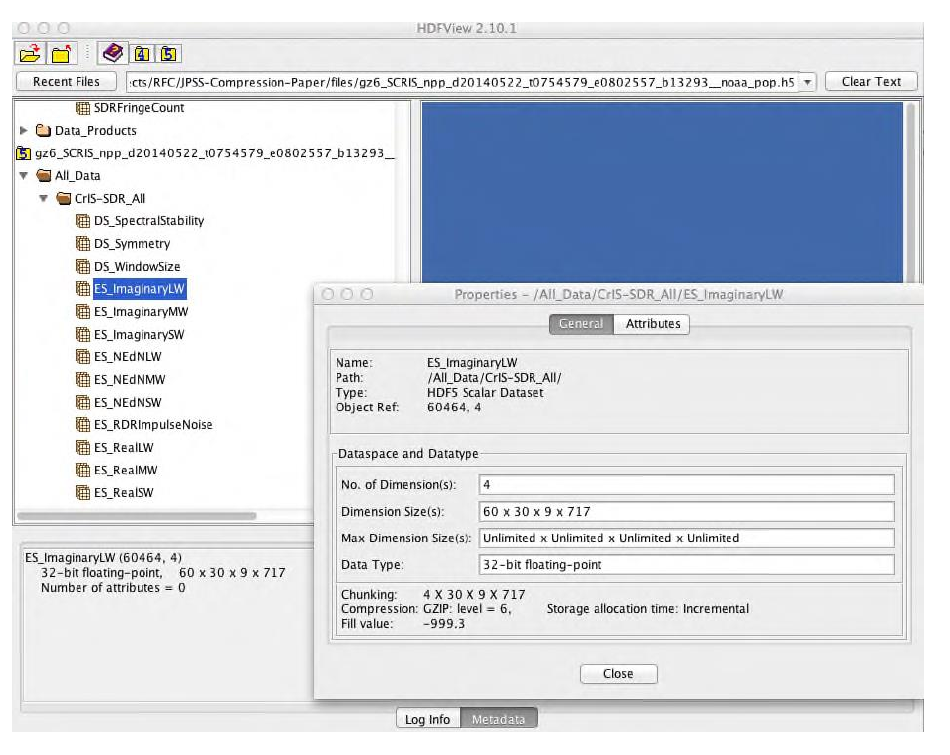
Figure 6 |
Our application read the dataset along the fastest changing dimension, 717 elements at a time from the dataset in both files. In the 2-dimensional case, this would correspond to reading an array by “row”. There were 16,200 reads to get all of the data. What we found was a several orders of magnitude drop in the performance when data was read from the compressed dataset as shown in Table 1.
| File Name | File.h5 | File_with_compression.h5 (gzip level 6) |
|---|---|---|
| Read Time | 0.1 seconds | 0.37 seconds |
We experimented with the HDF5 parameters such cache size and chunk size and modified our application to use different access patterns. The details of the experiments and achieved results will be discussed in the Tuning for Performance section. Here we provide the results just to show the difference in the read performance the change in the parameters made.
The table below, Table 2, shows the result of reading data as in the example above with the difference that the application used a chunked cache size of 3MB instead of the default 1MB. Reading performance from the compressed dataset was only 4 times slower than for reading the uncompressed data.
| File Name | File.h5 | File_with_compression.h5 (gzip level 6) |
|---|---|---|
| Read Time | 0.1 seconds | 0.37 seconds |
We also experimented with a different access pattern to read data from both files. Instead of reading 717 elements at a time, we read a contiguous HDF5 hyperslab with dimensions 4x30x9x717. The reader who knows about HDF5 chunking will immediately recognize that we read one chunk at a time, a total 15 of them. With this change, reading from the non-compressed dataset was only 10 times better than reading from the compressed dataset; see Table 3 below and compare with the results in Table 1.
| File Name | File.h5 | File_with_compression.h5 (gzip level 6) |
|---|---|---|
| Read Time | 0.04 seconds | 0.36 seconds |
In our last experiment, we repacked both files with h5repack to use a chunk size of 1x30x9x717, 4 times smaller than the original chunks, and read the file by using the original access pattern of 1x1x1x717 hyperslab (by “row”). The result is shown below in Table 4. Once again, we got much better performance than shown in Table 1, even when considering the time to repack the file with h5repack.
| File Name | File.h5 | File_with_compression-small-chunk.h5 (gzip level 6) |
|---|---|---|
| Read Time | 0.08 seconds | 0.36 seconds |
| Repack Time | 3 seconds | 12 seconds |
In this section we will give a brief overview of the chunking and compression features needed to follow the approach presented later in the “Tuning for Performance” section on page 15. For more information on HDF5 chunking, see the Chunking in HDF5 document.
Data of HDF5 dataset can be stored in several different ways in HDF5 file. See the Data Transfer Operations on a Dataset section in the HDF5 Datasets chapter in the HDF5 User Guide for more information.
The default storage layout of HDF5 files is contiguous storage: data of a multidimensional array is serialized (or flattened) along the fastest changing dimension and is stored as a contiguous block in the file. This storage mechanism is recommended if the size of a dataset is known and the storage size for the dataset is acceptable to the user: in other words, no data compression is desired. The contiguous storage is efficient for I/O if a whole HDF5 dataset is accessed or if a contiguous subset (as stored in the file) of an HDF5 dataset is accessed. The figure below shows an example with a row of a 2-dimensional array stored in an HDF5 dataset by a C application. In this case, the HDF5 Library seeks to the start position in the file and writes/reads the required number of bytes.
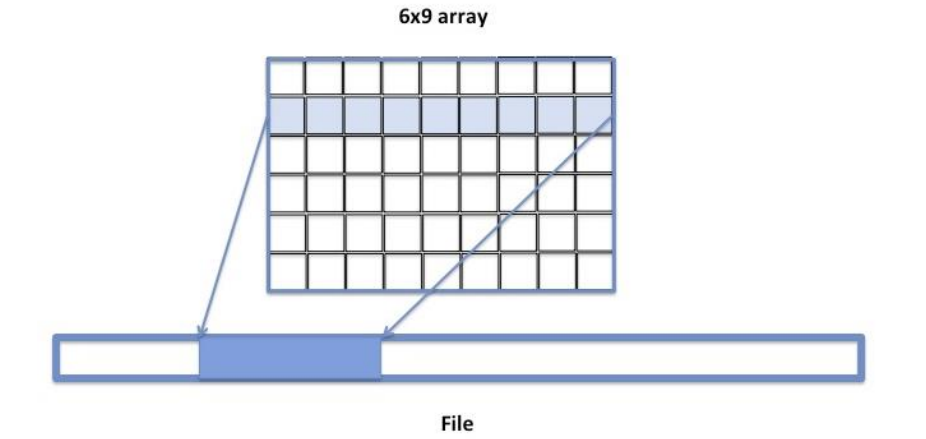
Figure 7 |
If we change the access pattern to accessing the dataset by columns instead of by rows, the contiguous layout may not work well. The column’s elements are not stored contiguously in the file (see Figure 8). Accessing a column will require several seeks to find the data in the file and multiple reads/writes of one element at a time. Seeks and small size I/O operations may affect performance especially for large datasets. Obviously, contiguous storage is not as favorable for a column access pattern as it is for a row access pattern, and other storage options may be more beneficial.
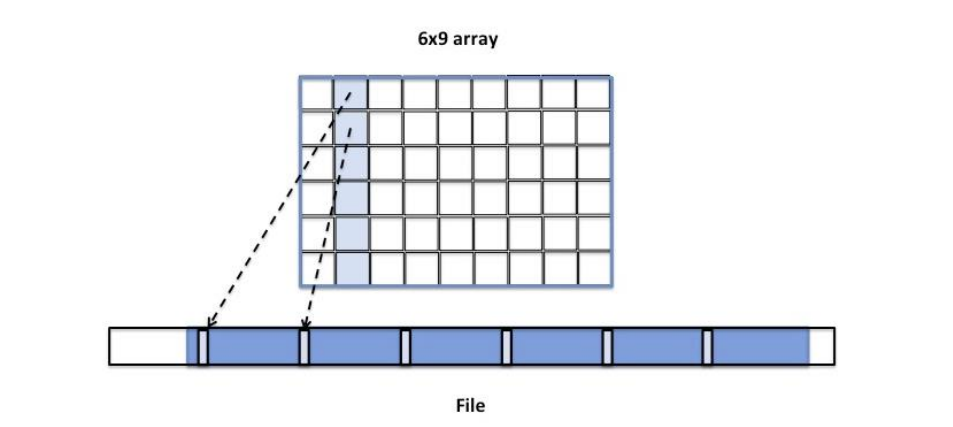
Figure 8 |
An alternative is chunked storage (a chunked storage layout). When chunked storage is used, a multidimensional array is logically divided into equally sized chunks. For example, Figure 9 below shows the 6x9 array divided into 6 3x3 chunks. Chunked storage layout and chunk sizes (number of elements in a chunk along each dataset dimension) are specified at dataset creation time and cannot be changed without rewriting the dataset. Chunked storage is required if data will be added to an HDF5 dataset and the maximum size of the dataset is unknown at creation time (see Figure 3). Chunked storage is also required if data will be stored compressed.
The logical chunk is stored as a contiguous block in the file (compare with the contiguous storage when the whole data array is stored contiguously in the file). When compression is used, it is applied to each chunk separately. During the I/O operation each chunk is accessed as a whole when the HDF5 Library reads or writes data elements stored in the chunk. For example, two chunks will be read (and uncompressed if needed) when accessing the 2nd column as shown in Figure 9.
The chunk size is an important factor in achieving good I/O and storage performance.
If the chunk size is too small, I/O performance degrades due to small reads/writes when a chunk is accessed. Storing a large number of small chunks increases the size of the internal HDF5 data structures needed to track the positions and sizes of chunks in the file, creating excessive storage overhead.
On the other hand, if the chunk size is too big and compression is used, I/O performance may degrade with unsuitable combinations of access patterns and chunk cache sizes or on systems that do not have enough memory to compress or to uncompress chunks. For instance, an application that reads data by row from a chunk too large to fit in the configured cache will cause decompression of the entire chunk for each row that is read, resulting in a great deal of unnecessarily repeated disk reads and decompression processing.
As was mentioned above, the storage layout cannot be changed after the dataset has been created. If desired, one can use the h5repack tool to modify the storage layout of a copy of a dataset; for example, the tool can be used to change the size of the chunk, to remove compression and store the dataset using contiguous storage, or to apply a different compression method. If data is read from the file many times, it may be much more efficient to rewrite the file using h5repack with the more appropriate storage parameters for reading, than to read data from the original file with an unfavorable compression and chunking arrangement.

Figure 9 |
Another important aspect of HDF5 chunking is the chunk cache.
HDF5 does not cache raw data unless chunked storage is used. When data is accessed for a chunked dataset, the chunks that contain the requested data are brought to the cache one by one and stay in cache until they are evicted. If a chunk is cached, then reading or writing data stored in the chunk does not require disk accesses. In other words, chunk caching helps when the same chunk is accessed multiple times during I/O operations.
The HDF5 Library provides the H5Pset_cache and H5Pset_chunk_cache functions to control the size of the chunk cache and the chunk eviction policy to specify the appropriate cache parameters for a particular access pattern.
As will be shown in the Tuning for Performance section, chunked storage and chunk cache parameters affect I/O performance and should be chosen with care depending on the I/O access pattern.
As it was mentioned in the previous sections, in HDF5 data can be stored compressed. The HDF5 Library comes with the built-in compression methods:
| H5Z_FILTER_DEFLATE | The gzip compression, or deflation, filter |
| H5Z_FILTER_SZIP | The SZIP compressionfilter |
| H5Z_FILTER_NBIT | The N-bit compression filter |
| H5Z_FILTER_SCALEOFFSET | The scale-offset compression filter |
| H5Z_FILTER_SHUFFLE | The shuffle algorithm filter |
| H5Z_FILTER_FLETCHER32 | The Fletcher32 checksum, or error checking, filter |
One can also build in a custom filter, Using HDF5 Filters, or use Using Dynamically-Loadable Filters.
The compression method is chosen at a dataset creation time and cannot be changed later. As with the chunked layout, one can use h5repack to rewrite the dataset in a copy of the dataset using a different compression method or to remove compression completely.
HDF5 tools such h5dump and h5ls can be used to check the efficiency of the compression. For example, both h5dump and h5ls show the compression ratio for a dataset. The compression ratio is defined as a ratio of the original data size to the size of compressed data. For example, the ratio for the dataset /All_Data/CrIS-SDR_All/ES_ImaginaryLW is 1.07 (see Figure 4) meaning that there was not much benefit in applying compression to save space in the file. For more information, see the HDF5 Compression Troubleshooting technical note for a discussion of compression efficiency.
The HDF5 Library applies compression encoding or decoding when the chunk is moved between the chunk cache and the file. Since compression encoding and decoding takes CPU time, it affects HDF5 write and read performance. This is especially true when data is read or written many times from the same chunk and the chunk is not cached between the accesses; this means the chunk has to be brought from disk every time it is accessed.
In the next section we will see the effect of compression on the I/O performance.
In this section we will discuss several strategies one can apply to get better I/O performance. We will explain in detail how a particular strategy works and when it should be applied. While the examples below focus on reading only, the same approach will work for writing too.
The strategies for improving performance require modifications to the reading application or to the HDF5 file itself. The reader should choose the strategies that are appropriate for a particular use case.
The HDF5 Library automatically creates a chunk cache for each opened chunked dataset. The first strategy is to check whether the current chunk cache settings work properly with the application access pattern and reset the chunk cache parameters as appropriate.
The HDF5 Library provides two functions, H5Pset_cache and H5Pset_chunk_cache, to control chunk cache settings. H5Pset_cache controls the chunk cache setting for ALL datasets in the file, and H5Pset_chunk_cache controls the chunk cache settings per dataset. To find out the default or current settings, use the H5Pget_cache or H5Pget_chunk_cache functions and then reset appropriate parameters if necessary. See the How to Adjust the Chunk Cache Size section for more information.
The default size of the cache is 1MB. The size can be modified by setting the nbytes parameter in H5Pset_cache and H5Pset_chunk_cache. Several chunks can be held in the cache if their total size in bytes is less or equal to 1MB.
To look up a chunk in cache, the HDF5 Library uses an array of pointers to the chunks (hash table). The array has nslots elements (or slots in the hash table) with a default value of 511. One can use the nslots parameter in H5Pset_cache and H5Pset_chunk_cache to change the default size of the hash table.
Each chunk has an associated hash value that is calculated as follows. All chunks of the dataset have an index (cindex) in a linear array of chunks. For example, chunks in Figure 9 will have indices from 0 to 5, with the upper left chunk having index 0, the middle one in the top row having index 1, and the lower right chunk having index 5. The hash value is calculated as the remainder of dividing cindex by nslots (known as a modulo operation cindex mod nslots). The hash table can contain only one chunk with the same hash value. This fact is important to remember to avoid situations when the needed chunks have the same hash value. For example, let’s assume nslots is 3. Then in Figure 9 the chunks with the indices 0 and 3 (in other words, the chunks that contain the first three columns) have the same hash values and cannot be in the chunk cache simultaneously even though their total sizes are less than 1MB.
Now, we can analyze what happens when data is read by “rows” (contiguous 717 elements) from the /All_Data/CrIS-SDR_All/ES_ImaginaryLW dataset and the default chunk cache settings are used. The number of slots nslots in the hash table is not a concern since the default value is 511 and we have only 15 chunks. Now let’s analyze how the chunk cache size affects the performance.
Each row is stored in one of the 15 chunks that comprise the dataset. Each chunk has 4x30x9 or 1,080 “rows”. To read the first row of the chunk, the whole chunk is read, uncompressed and the row is copied to the application buffer by the HDF5 Library. Since the size of the uncompressed chunk is 2.95 MB, the cache cannot hold the chunk. When the second row is read, the process repeats until all rows from the same chunk are read. Thus, the chunk will be read and uncompressed 1,080 times. When we increase the cache size to 3MB, the chunk stays in the cache and all rows can be copied to the application buffer without the HDF5 Library fetching data from disk and uncompressing the chunk every time the chunk is accessed.
Since all 15 chunks have to be read, the HDF5 Library will be touching the disk 16,200 times when a 1MB size cache is used compared with 15 times when a 3MB cache is used. The first column in Table 5 below shows that it took 345 seconds to read a compressed dataset when using the default cache size of 1MB while it took only 0.37 seconds to read the dataset when using the chunk cache size of 3MB. We see several orders of magnitude performance improvements when we increase chunk cache size to 3MB.
| File Name | File_with_compression.h5 | File_with_compression.h5 |
|---|---|---|
| Cache Size | 1MB (default) | 3MB |
| Read Time | 345 seconds | 0.37 seconds |
As shown in Table 6 below, the reading performance with the 3MB cache size is comparable to the reading performance of the data stored without compression applied. Please notice that the chunk cache size did not affect the reading performance for the uncompressed data.
| File Name | File_with_compression.h5 | File.h5 |
|---|---|---|
| Cache Size | 3MB | 1MB or 3MB |
| Read Time | 0.37 seconds | 0.1 seconds |
As was mentioned above, an application can adjust the chunk cache size by calling either H5Pset_cache or H5Pset_chunk_cache functions. H5Pset_cache sets the chunk cache size for all chunked datasets in a file, and H5Pset_chunk_cache sets the chunk cache size for a particular dataset.
The programming model for using both functions is the following:
Below are the code snippets that show the usage.
The first example below shows how to change the cache size for all datasets in the file using H5Pset_cache. Since the function sets a global setting for the file, it uses a file access property list identifier to modify the cache size. H5Pget_cache is called first to retrieve default cache settings that will be modified by H5Pset_cache. In the example below, every chunked dataset will have a cache size of 3MB. To overwrite this setting for a particular dataset one can use H5Pset_chunk_cache as shown in the second example.
| Code Example 1: Using H5Pset_cache to change the cache size for all datasets. |
|---|
The second example, see below, shows how to set at dataset creation time the chunk cache size for the /All_Data/CrIS-SDR_All/ES_ImaginaryLW dataset. The cache sizes for other datasets will not be modified.
| Code Example 2 : Using H5Pset_chunk_cache to change one dataset. |
|---|
As we will see in the next section, care needs to be taken when working with chunked datasets and setting chunk cache sizes: an application’s memory footprint can be significantly affected.
A chunk cache is allocated for a dataset when the first I/O operation is performed. The chunk cache is discarded after the dataset is closed. If an application performs I/O on several datasets, memory consumed by an application increases by the total size of all chunk caches. One can also see an increase in the metadata cache size.
If memory consumption is a concern, it is recommended that I/O be done on a few datasets at a time and to close the few datasets after I/O operation has been completed. As we will see in the next sections, there are access patterns that cannot take advantage of a chunk cache at all. If this is the case, the application can disable a chunk cache completely and thus reduce the memory footprint. To disable a chunk cache, use 0 for the value of the nbytes parameter in the calls to H5Pset_cache or H5Pset_chunk_cache.
When changing the chunk cache size is not an option (for example, there is no access to the program source code), one can consider a reading strategy that will minimize the effect of the chunk cache size. The strategy is to read as much data as possible in each read operation.
As we mentioned before, the HDF5 Library performs I/O on the whole chunk. The chunk is read, uncompressed, and the requested data is copied to the application buffer. If in one read call the application requests all data in a chunk, then obviously chunk caching (and chunk cache size) is irrelevant since there is no need to access the same chunk again.
In our case, suppose the application reads the selection that corresponds to the whole chunk. In other words, if a hyperslab with dimensions 4x30x9x717 is used instead of a hyperslab with dimensions 1x1x1x717, then the HDF5 Library would perform only 15 reading and decoding operations instead of 16,200. The significant improvement in performance is shown in Table 7 below. We see a similar I/O performance improvement as in the case when we increased the chunk cache size to 3MB (see Table 5).
| File Name | File_with_compression.h5 | File_with_compression.h5 |
|---|---|---|
| Access Pattern | 1x1x1x717 | 4x30x9x717 |
| Read Time | 345 seconds | 0.36 seconds |
Data producers should consider that users who cannot modify applications to increase the chunk cache size or to change the access pattern will not encounter the performance problem described in the How to Adjust the Chunk Cache Size section if chunks in the file are smaller than 1MB (1x30x9x717 by 4 bytes) because the whole chunk will fit into the chunk cache of the default size. Therefore if data in the HDF5 files is intended for reading by unknown user applications or on systems that might be different from the system where it was written, it is a good idea to consider a chunk size less than 1MB. In this case the applications that use default HDF5 settings will not be penalized.
As shown in the Case Study section, Table 4, the performance of reading by row (717 elements) when the chunk size is 1x30x9x717 (total size in bytes is approximately 0.74MB) is comparable to the performance of reading non-compressed data and is similar to the performance for reading compressed data when using a bigger cache size (Table 2) or bigger amount of data (Table 3). The above statement is summarized in the Recommendations section.
For users who encounter datasets with large chunk sizes and with applications that cannot be easily modified: since the chunk size is set at the dataset creation time and cannot be changed later, the only option is to recreate the dataset by using the h5repack tool to change the storage layout properties. The command below will change the chunk size of the /All_Data/CrIS-SDR_All/ES_ImaginaryLW dataset from 4x30x9x717 to 1x30x9x717 making chunk size in bytes 0.74MB instead of the original 2.96MBs size.
This section summarizes the discussion and recommendations for working with files that use the HDF5 chunking and compression feature.
When compression is enabled for an HDF5 dataset, the library must always read an entire chunk for each call to H5Dread unless the chunk is already in the cache. To avoid trashing the cache, make sure that the chunk cache size is big enough to hold the whole chunk or that the application reads the whole chunk in one read operation bypassing the chunk cache.
When experiencing I/O performance problems with compressed data, find the size of the chunk and try the strategy that is most applicable to your use case:
The results of all three strategies provide similar performance and are summarized in Table 8 below.
| File Name | File_with_compression.h5 | File_with_compression.h5 | File_with_compression.h5 | File_with_compression-small-chunk.h5 |
|---|---|---|---|---|
| Cache Size | 1MB | 3MB | 1MB | 1MB |
| Chunk Size | 4x30x9x717 | 4x30x9x717 | 4x30x9x717 | 1x30x9x717 |
| Access Pattern (Hyperslab Size) | 1x1x1x717 | 1x1x1x717 | 4x30x9x717 | 1x1x1x717 |
| Read Time | 345 seconds | 0.37 seconds | 0.36 seconds | 0.36 seconds |
| Repack Time | NA | NA | NA | 12 seconds |
Please notice that when compression is disabled, the library’s behavior depends on the cache size relative to the chunk size. If the chunk fits the cache, the library reads entire chunk for each call to H5Dread unless it is in cache already. If the chunk doesn’t fit the cache, the library reads only the data that is selected directly from the file. There will be more read operations, especially if the read plane does not include the fastest changing dimension.
One can use h5repack tool to remove compression by using the following command:
The CCP tool described in the introduction is intended to facilitate optimization of the parameters chosen when creating files and investigation of possible solutions when performance problems are encountered.
Navigate back: Main / Technical Notes